How intracellular signaling regulates growth factor production
Cancer cells need life-essential molecules to proliferate. These so-called growth factors are activated by ectodomain shedding of precursor proteins on the outside of the plasma membrane, mainly carried out by three human cleavage enzymes. A pharmaceutical blocking of these enzymes could hinder cancer from growing but would also inhibit other life-essential processes. Now, researchers from German Leibniz Institute for Age Research (FLI) and Harvard University, US, showed that the factor-precursor-producing cells themselves determine if and when cleavage may occur. This is decided by intracellular signaling. Interfering with defined signaling in cells producing cancer growth factors could be developed into a new way of cancer treatment.
As cancer cells proliferate in an unlimited way, they need to be supplied with oxygen and nutrients. For their growth and the formation of blood vessels, so-called growth factors are required. These hormone-like proteins are activated by the shedding of transmembrane precursor proteins that have to be cleaved on the outside of the plasma membrane by specialized enzymes. In the human body, mainly three "cleavage enzymes" are responsible for ectodomain shedding of hundreds of growth factors. Hindering one of these enzymes from cleaving would certainly suppress the production of growth factors related to tumorigenesis, but would have severe side-effects: a lot of life-essential molecules would also be inhibited. Since ectodomain cleavage is highly important for homeostasis of the organism, it needs to be tightly regulated with respect to both its overall abundance and time course.
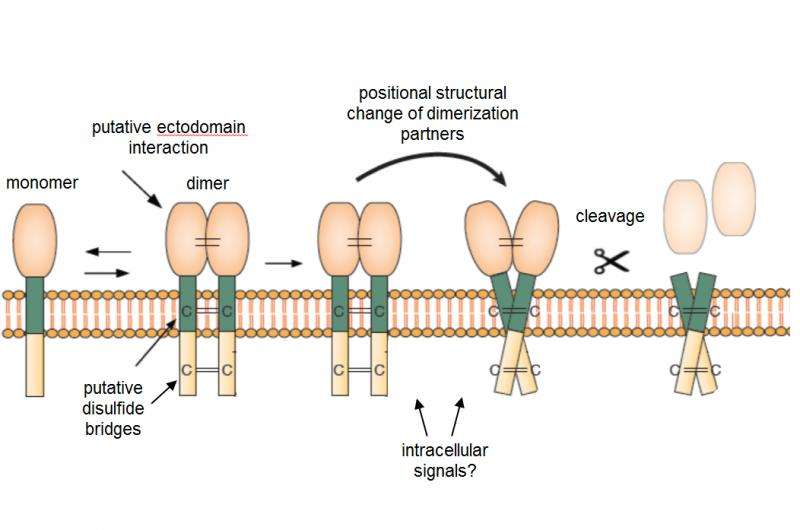
Now, in a collaborative project, researchers from German Leibniz Institute for Age Research (FLI) in Jena and renowned Harvard University in Cambridge, US, showed that obviously the precursor proteins themselves dictate if and when the "scissor"-enzymes may cut. Signal processing in the intracellular domain of the precursor-protein-producing cells is responsible for modifications that likely induce a relative positional change of the dimerization partners and, in the end, allow cleavage. This is individually different for each precursor protein. The collaborators from Jena and Cambridge already found many details of the mechanism to explain how the intracellular domain modification communicates with the ectodomain of the substrate to allow for cleavage to occur, e.g. releasing growth factors linked to breast cancer (Epidermal Growth Factor family) and Neuregulin which is important for neuro-regeneration, as well as cleavage of a protein relevant for metastasizing of cancer cells. The latest publication in the Journal of Biological Chemistry now was nominated as one of the best 50 out of this year's 6.000 publications.
"Our research results offer a new way of suppressing growth factors related to cancer cell proliferation", Prof. Dr. Peter Herrlich, former scientific director and now associated researcher at FLI, explains. Instead of blocking the cleavage enzymes and condoning side-effects, the intracellular signal processing for single precursor proteins may be inhibited in order to specifically knock out the growth factors required by individual cancer types.
More information: "Inside-out Regulation of Ectodomain Cleavage of Cluster-of-Differentiation-44 (CD44) and of Neuregulin-1 Requires Substrate Dimerization." Journal of Biological Chemistry (2015), DOI: 10.1074/jbc.M114.610204
Journal information: Journal of Biological Chemistry
Provided by Fritz Lipmann Institute