May 28, 2015 report
Self-replicating nanostructures made from DNA
(Phys.org)—Is it possible to engineer self-replicating nanomaterials? It could be if we borrow nature's building blocks. DNA is a self-replicating molecule where its component parts, nucleotides, have specific chemical interactions that allow for the design of self-assembled structures. In biological systems, DNA replicates with the aid of proteins. However, Junghoon Kim, Junwye Lee, Shogo Hamada, Satoshi Murata, and Sung Ha Park of Sungkyunkwan University and Tohoku University have designed a controllable self-replicating system that does not require proteins. Their work appears in Nature Nanotechnology.
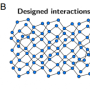
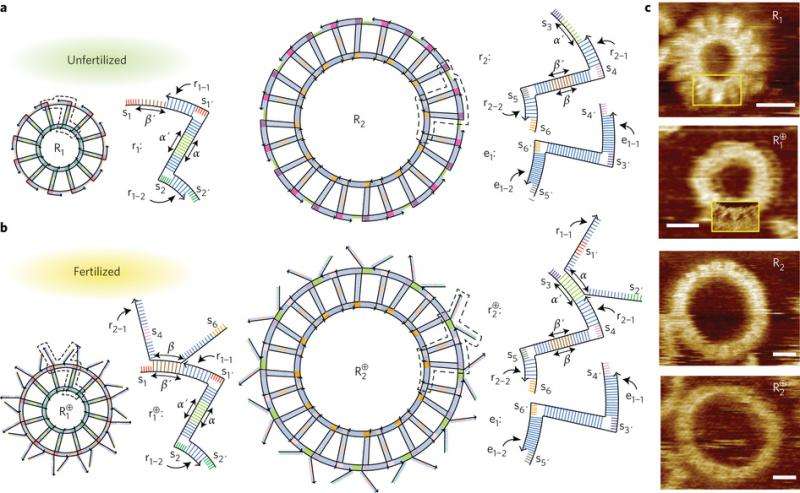
To understand how this self-replicating process works, it is important to know the different component parts. Kim et al. designed two DNA T-motifs, r1 and r2, which are double-stranded DNA comprised of functional domains, labeled alpha and beta, and "sticky" ends as connection points. They also designed an extension motif. Twelve units of the r1 motif self-assemble into a small ring, R1, and twelve units of r2 plus twelve extension motifs self-assemble into a larger ring, R2.
These components can be in two different states, "fertilized" or "unfertilized". The fertilized structures contain the features necessary for replication. Fertilization happens when a single-stranded alpha or beta domain of an r1 or r2 motif binds with a strand having a complementary alpha or beta domain. This leaves a single-strand protrusion, or toehold, extending from the ring or from the original motif. The toeholds indicate that the ring or motif is fertilized.
These toeholds extending from the DNA ring bind to complimentary invader strands. When this happens, the hybridized structure consisting of the toehold and the invading strand breaks off of the initial ring, and eventually, as these pieces break off due to branch migration, they self-assemble into another ring.
This process continues through two different replication pathways. One pathway grows exponentially. The other pathway grows according to Fibonacci's sequence. The particular pathway taken depends on which invading strands are added to the system.
The authors verified that the DNA ring populations grew through this toehold-mediated process with AFM and absorbance studies. For the AFM studies, they took a small sample from each phase and determined the average number of rings present in that phase. Absorbance data was adjusted to determine the relative concentration of rings at each phase.
They also verified that the daughter rings were a result of annealing to the single-strand toeholds from the initial ring rather than as a result of self-assembly of residual DNA motifs in solution using gel electrophoresis and extracting the DNA products from each phase. The individual phases were studied with AFM and invading strands were added to a solution during each of the phases to see if rings formed.
Kim, et al. demonstrated that nanoscale self-replication can occur using the thermodynamic properties of toehold-mediated strand displacement and the self-assembly abilities of DNA motifs. In this study synthetic DNA T-motifs self-assemble into structures that allow for sequential reactions to take place. This research demonstrates the possibility of functionally programmable self-replicating nanostructures.
More information: "Self-replication of DNA rings" Nature Nanotechnology, DOI: 10.1038/nnano.2015.87
Abstract
Biology provides numerous examples of self-replicating machines, but artificially engineering such complex systems remains a formidable challenge. In particular, although simple artificial self-replicating systems including wooden blocks, magnetic systems, modular robots and synthetic molecular systems have been devised, such kinematic self-replicators are rare compared with examples of theoretical cellular self-replication. One of the principal reasons for this is the amount of complexity that arises when you try to incorporate self-replication into a physical medium. In this regard, DNA is a prime candidate material for constructing self-replicating systems due to its ability to self-assemble through molecular recognition. Here, we show that DNA T-motifs, which self-assemble into ring structures, can be designed to self-replicate through toehold-mediated strand displacement reactions. The inherent design of these rings allows the population dynamics of the systems to be controlled. We also analyse the replication scheme within a universal framework of self-replication and derive a quantitative metric of the self-replicability of the rings.
Journal information: Nature Nanotechnology
© 2015 Phys.org