May 19, 2015 report
General principles to explain DNA brick self-assembly
(Phys.org)—DNA bricks are an odd phenomenon. They are nanostructures built from synthetic, single-strand DNA that self-assemble into 3D structures. Several years ago, researchers at Wyss Institute at Harvard demonstrated the self-assembly of designed 3D structures made from 1,000 "bricks" that fit together like molecular Legos. One of the curiosities about these DNA bricks is why this complex self-assembly process works so well.
In an effort to uncover the nuances of DNA brick self-assembly, William M. Jacobs, Aleks Reinhardt, and Daan Frenkel from the University of Cambridge conducted theoretical studies to understand what drives the assembly process. Their studies reveal how temperature, kinetics, coordination number, and bond energies, contribute to the unique behavior of DNA brick self-assembly, and while it is a nucleated process, it does not behave according to classical nucleation theory. Their work appears in the Proceedings of the National Academy of Sciences.
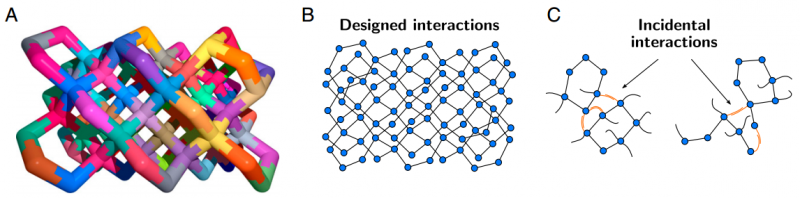
In order to optimize DNA brick assembly, Jacobs, et al. wanted to investigate whether these systems had a nucleation barrier. In classic nucleation theory (CNT), the nucleation barrier is the energy cost for making an ordered structure, such as a crystal. This energy barrier must be overcome before a compound can settle into its equilibrium structure. The authors found that DNA brick assembly does have a nucleation barrier, but, unlike CNT, this barrier is determined by the way the bricks are connected in the target structure. They identified several factors that contribute to this non-classical behavior.
DNA bricks are designed to assemble in particular ways by matching certain sections of a DNA strand with a complimentary section. These designed interactions need to be stronger than incidental interactions. They found that there is a critical number of DNA strands that must be assembled to promote the formation of the target structure, meaning that the the nucleation barrier is a consequence of the number of subunits that must be assembled.
Additionally, DNA-bricks have a certain temperature range at which the nucleation barrier is overcome, but over which the free energy of the final structure is higher than the initial monomers. This deviates from CNT which predicts that a nucleus with more than the critical number of DNA strands will assemble into the complete structure. This difference is due to DNA bricks forming stable intermediate structures. Experimentally, this means that self-assembly occurs in a narrow temperature range in which nucleation begins at a high temperature, and as the structure is gradually cooled, the target structure is favored.
In prior studies, the maximum number of bonds made by each brick in the interior of a structure was four, but it is possible to have different coordination numbers. The question was whether the coordination number affected the nucleation barrier, and whether four bonds were necessary. It turns out that DNA brick self-assembly works well because each subunit makes only a small number of bonds. When the authors tested higher coordination numbers, they found that the system behaved analogously to CNT.
Finally, Jacobs et al. found that contrary to theories that homogenous bond energies would decrease the number of errors during DNA brick assembly, it is the diversity in bond energies that contributes to enhanced kinetics of DNA brick self-assembly. This was likely due to stabilizing unstable intermediate structures.
Jacobs, et al. have identified several key features that aid in understanding why DNA brick self-assembly works as well as it does. Features such as ramping down the temperature and optimizing the coordination number explain why there are so few errors in the self-assembly process. And, their discovery that heterogeneous bond energies promote target molecule formation is a practical place where researchers can improve yields and decrease errors in DNA brick assembly.
More information: "Rational design of self-assembly pathways for complex multicomponent structures" PNAS, DOI: 10.1073/pnas.1502210112
Abstract
The field of complex self-assembly is moving toward the design of multiparticle structures consisting of thousands of distinct building blocks. To exploit the potential benefits of structures with such "addressable complexity," we need to understand the factors that optimize the yield and the kinetics of self-assembly. Here we use a simple theoretical method to explain the key features responsible for the unexpected success of DNA-brick experiments, which are currently the only demonstration of reliable self-assembly with such a large number of components. Simulations confirm that our theory accurately predicts the narrow temperature window in which error-free assembly can occur. Even more strikingly, our theory predicts that correct assembly of the complete structure may require a time-dependent experimental protocol. Furthermore, we predict that low coordination numbers result in nonclassical nucleation behavior, which we find to be essential for achieving optimal nucleation kinetics under mild growth conditions. We also show that, rather surprisingly, the use of heterogeneous bond energies improves the nucleation kinetics and in fact appears to be necessary for assembling certain intricate 3D structures. This observation makes it possible to sculpt nucleation pathways by tuning the distribution of interaction strengths. These insights not only suggest how to improve the design of structures based on DNA bricks, but also point the way toward the creation of a much wider class of chemical or colloidal structures with addressable complexity.
Journal information: Proceedings of the National Academy of Sciences
© 2015 Phys.org