April 2, 2015 report
Using DNA to assemble a protein lattice

(Phys.org)—Living systems have ready-made catalysts, known as enzymes, for many types of reactions. What if these enzymes could be snapped together like Lego pieces to make a lattice structure? An ideal procedure for accomplishing this would be versatile enough to work with any protein, regardless of its identity, and would maintain the protein's native functionality while in the lattice structure.
One group has accomplished a proof-of-concept experiment for making a protein superlattice. Jeffrey D. Brodin and Evelyn Auyeung, working with Professor Chad A. Mirkin at Northwestern University have devised a general strategy for making a protein lattice out of one or two different proteins and out of a combination of proteins and gold nanoparticles. Their research is reported in The Proceedings of the National Academy of Sciences.
Dr. Mirkin of Northwestern University spearheaded research on spherical nucleic acids (SNAs), a form of DNA his group discovered in 1996, in which a nanoparticle is functionalized with DNA via a covalent bond. The DNA creates a "shell" around the nanoparticle and displays unique properties compared to its linear counterpart. In this current research, Dr. Mirkin's group developed a new strategy for functionalizing enzymes with DNA.
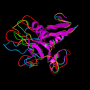
For this proof-of-concept procedure, they used two well-known catalases, bovine catalase and corynebacterium glutamicum (Cg) catalase, to serve as their core particle, analogous to the gold nanoparticle in their SNA gold nanoparticle structures. These proteins catalyze hydrogen peroxide (H2O2) decomposition with a known kinetic profile, so they serve as good model systems for comparison studies.
The first step was to attach oligonucleotides onto the surface of the catalases. Both catalases have surface-accessible amines that were converted to azides using a procedure that was optimized to preserve the structure and function of the protein while maximizing the number of functional amines. Oligonucleotides were then added to the azide-modified catalases using copper-free click chemistry, a biologically inert cycloaddition procedure.
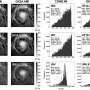
Ultraviolet-Visible (UV-Vis) spectroscopy showed that the density of oligonucleotides on the catalases was 30 to 50 pmol/cm2. Dynamic light scattering confirmed a shell-like structure of oligonucleotides radiating from the protein core, similar to its nanoparticle counterparts. Characterization with UV-Vis and circular dichroism confirmed that the functionalized catalases maintained their native structures. Reaction studies, monitored with UV-Vis, confirmed that the functionalized catalases had an intact active site that maintained its ability to catalyze H2O2 decomposition.
The next step was to see if these functionalized proteins formed lattices similar to the SNA gold nanoparticle body-centered cubic lattice structure. They tested two different types of systems. One system contained both of the functionalized catalases in a 1:1 ratio. The other system had a functionalized catalase and spherical nucleic acids with a gold nanoparticle core similar in size to the catalase.
To promote lattice formation, the systems were heated to above the melting point, but below the point of denaturing the catalase. They were allowed to cool at a specific rate that was determined in prior studies in which crystallization was optimized. X-ray scattering revealed that all four possible systems (Cg catalase, Cg and bovine catalase, Cg and functionalized gold nanoparticles, and bovine and functionalized gold nanoparticles) showed CsCl-like lattice packing. While the lattices composed of proteins and gold nanoparticles showed the expected scattering patterns, the CsCl-like packing of lattices composed of identical proteins suggests that each catalase is surrounded by eight neighbors, but that the surface chemistry as well as the shape of the catalase impose distinct orientations of proteins at each lattice position.
Importantly, lattice formation was not based on the catalase's chemical make-up or other non-specific interactions, but on complementary oligonucleotides directing the assembly of the catalases into lattice structures. The final step was to confirm that even in a lattice structure, the catalases maintained their functionality. The Cg lattice was used to catalyze the decomposition of H2O2. Kinetic studies showed that the reaction was still first-order with respect to H2O2 concentration, but that the reaction was slower by a factor of 20. This is most likely due to the molecular diffusion into the lattice structure. Furthermore, the catalytic lattice was recoverable using centrifugation and it was reusable in subsequent reactions.
Overall, this procedure provides a viable method for producing lattice structures comprised of various types of enzymes. It also provides a potential mechanism for influencing lattice structure by choosing proteins with particular symmetries or arrangements of functionalizable amino acids. Additionally, one could construct lattice structures from nanoparticles and proteins. Future research might explore bio-engineering applications.
More information: "DNA-Mediated engineering of multicomponent enzyme crystals" Jeffrey D. Brodin, PNAS, DOI: 10.1073/pnas.1503533112
Journal information: Proceedings of the National Academy of Sciences
© 2015 Phys.org