Squid recode their genetic make-up on-the-fly to adjust to their surroundings

The principle of adaptation—the gradual modification of a species' structures and features—is one of the pillars of evolution. While there exists ample evidence to support the slow, ongoing process that alters the genetic makeup of a species, scientists could only suspect that there were also organisms capable of transforming themselves ad hoc to adjust to changing conditions.
Now a new study published in eLife by Dr. Eli Eisenberg of Tel Aviv University's Department of Physics and Sagol School of Neuroscience, in collaboration with Dr. Joshua J. Rosenthal of the University of Puerto Rico, showcases the first example of an animal editing its own genetic makeup on-the-fly to modify most of its proteins, enabling adjustments to its immediate surroundings. The research, conducted in part by TAU graduate student Shahar Alon, explored RNA editing in the Doryteuthis pealieii squid.
"We have demonstrated that RNA editing is a major player in genetic information processing rather than an exception to the rule," said Dr. Eisenberg. "By showing that the squid's RNA-editing dramatically reshaped its entire proteome—the entire set of proteins expressed by a genome, cell, tissue, or organism at a certain time—we proved that an organism's self-editing of mRNA is a critical evolutionary and adaptive force." This demonstration, he said, may have implications for human diseases as well.
Using the genetic red pencil
RNA is a copy of the genetic code that is translated into protein. But the RNA "transcript" can be edited before being translated into protein, paving the way for different versions of proteins. Abnormal RNA editing in humans has been observed in patients with neurological diseases. The changing physiological appearance of squid and octopuses over their lifetime and across different habitats has suggested extensive recoding might occur in these species. However, this could never be confirmed, as their genomes (and those of most species) have never been sequenced.
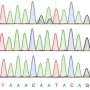
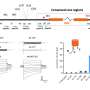
For the purpose of the new study, the researchers extracted both DNA and RNA from squid. Harnessing DNA sequencing and computational analyses at TAU, the team compared the RNA and DNA sequences to observe differences. The sequences in which the RNA and DNA did not match up were identified as "edited."
"It was astonishing to find that 60 percent of the squid RNA transcripts were edited. The fruit fly, for the sake of comparison, is thought to edit only 3% of its makeup," said Dr. Eisenberg. "Why do squid edit to such an extent? One theory is that they have an extremely complex nervous system, exhibiting behavioral sophistication unusual for invertebrates. They may also utilize this mechanism to respond to changing temperatures and other environmental parameters."
"Misfolding" the proteins
The researchers hope to use this approach to identify recoding sites in other organisms whose genomes have not been sequenced.
"We would like to understand better how prevalent this phenomenon is in the animal world. How is it regulated? How is it exploited to confer adaptability?" said Dr. Eisenberg. "There may be implications for us as well. Human diseases are often the result of 'misfolded' proteins, which often become toxic. Therefore the question of treating the misfolded proteins, likely to be generated by such an extensive recoding as exhibited in the squid cells, is very important for future therapeutic approaches. Does the squid have some mechanism we can learn from?"
Journal information: eLife
Provided by Tel Aviv University