A faster way to flag bacteria-tainted food and prevent illness
The regular appearance of food poisoning in the news, including a recent event that led to the recall of more than 33,000 pounds of chicken, drives home the need for better bacterial detection long before meats and produce make it to the dinner table. On the horizon is a new approach for pathogen screening that is far faster than current commercial methods. Scientists are reporting the technique in the ACS journal Analytical Chemistry.
Sibani Lisa Biswal and colleagues note that Salmonella is one of the pathogens most commonly associated with foodborne illness, which can cause fever, diarrhea and abdominal cramps. An estimated one in six Americans suffer from food poisoning every year, according to the Centers for Disease Control and Prevention. Many end up in the hospital, and about 3,000 people die annually. Conventional methods to detect harmful bacteria in food are reliable and inexpensive, but they can be complicated, time consuming and thus allow contamination to go undetected. Biswal's team set out to develop a faster method to catch unwanted microbes before they can make people sick.
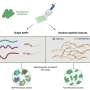
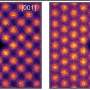
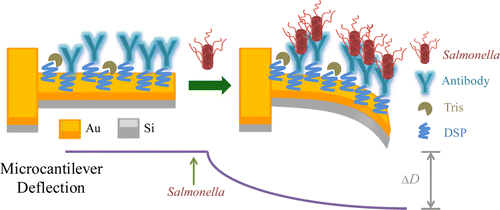
They used an array of tiny "nanomechanical cantilevers," anchored at one end, kind of like little diving boards. The cantilevers have peptides attached to them that bind to Salmonella. When the bacteria bind to the peptides, the cantilever arm bends, creating a signal. The screening system rapidly distinguished Salmonella from other types of bacteria in a sample. One of the peptides was even more specific than an antibody, which is considered the gold standard. That peptide could tell eight different types of Salmonella apart from each other. The researchers stated that the technique could be applied to other common food pathogens.
More information: "Rapid Detection of Pathogenic Bacteria and Screening of Phage-Derived Peptides using Microcantilevers" Anal. Chem., Article ASAP, DOI: 10.1021/ac403437x
Abstract
We report the use of an array of microcantilevers to measure the specific binding of Salmonella to peptides derived from phage display libraries. Selectivity of these phage-derived peptides for Salmonella spp. and other pathogens (Listeria monocytogenes and Escherichia coli) are compared with a commercially available anti-Salmonella antibody and the antimicrobial peptide alamethicin. A Langmuir isotherm model was applied to determine the binding affinity constants of the peptides to the pathogens. One particular peptide, MSal 020417, demonstrated a higher binding affinity to Salmonella spp. than the commercially available antibody and is able to distinguish among eight Salmonella serovars on a microcantilever. A multiplexed screening system to quickly determine the binding affinities of various peptides to a particular pathogen highly improves the efficiency of the peptide screening process. Combined with phage-derived peptides, this microcantilever-based technique provides a novel biosensor to rapidly and accurately detect pathogens and holds potential to be further developed as a screening method to identify pathogen-specific recognition elements.
Journal information: Analytical Chemistry
Provided by American Chemical Society