Research describes new techniques to study protein-DNA interactions
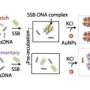
Work undertaken at the John Innes Centre describes new Surface Plasmon Resonance (SPR) protocols to identify and footprint protein-DNA interactions in a cost effective and semi-automated way.
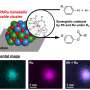
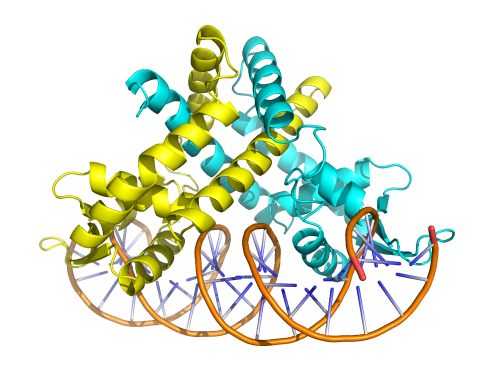
This work, published in Nucleic Acids Research, illustrates the application of these methods by locating specific binding sites for a bacterial transcription factor and accurately defining the protein footprints on the DNA. These observations were then verified by determining the structure of a representative protein-DNA complex using X-ray crystallography.
These new methods will appeal to those investigating protein-nucleic acid interactions as an alternative to traditional methods, such as electrophoretic mobility shift assays and DNase I footprinting.
The SPR protocols are based on an indirect DNA capture method that has been termed ReDCaT (Re-usable DNA Capture Technique). ReDCaT is now being offered as a service by Inspiralis, a company founded in 1995 by John Innes Centre researchers.
More information: Stevenson, C. et al. (2013). Investigation of DNA sequence recognition by a streptomycete MarR family transcriptional regulator through surface plasmon resonance and X-ray crystallography, Nucleic Acids Research. nar.oxfordjournals.org/content … 6/07/nar.gkt523.full
Journal information: Nucleic Acids Research
Provided by John Innes Centre