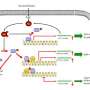
Pairs of gene-regulating proteins work together to identify their proper binding sites within the genome
Research led by Lawrence Stanton and Prasanna Kolatkar of the A*STAR Genome Institute of Singapore has provided valuable insight into embryonic development. Stanton and Kolatkar teamed up with colleagues in Singapore to determine how transcription factor proteins assemble at DNA sequences called enhancers, which help coordinate gene expression.
Multiple transcription factors often act together at a given enhancer, but biologists are unsure whether these proteins assemble sequentially at the enhancer or combine by preforming complexes that collaboratively recognize their targets.
The researchers focused on a transcription factor called Oct4, which is known to help embryonic stem cells maintain a 'pluripotent' state, from which they can give rise to any tissue in the body. However, Oct4 also helps direct the development of endodermal tissue, which forms the nervous system and other tissues. Stanton and Kolatkar suspected that Oct4's effects might be determined by the transcription factors with which it partners.
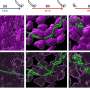
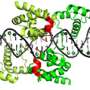
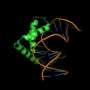
Focusing on two particular transcription factors, Sox2 and Sox17, the team identified different sets of enhancers that are selectively targeted by Sox2/Oct4 versus Sox17/Oct4. Interestingly, the binding sequences differed subtly for the two transcription factor pairs: Sox17/Oct4 preferentially bound a 'compressed' motif that was one base pair shorter than the 'canonical' motif typically bound by Sox2/Oct4. A series of experiments with mouse embryonic stem cells showed that gene enhancers associated with endoderm formation tend to contain the compressed motif and are therefore switched on by Sox17/Oct4 (see image). Conversely, Sox2/Oct4 preferentially gravitates to pluripotency-inducing enhancers containing the canonical motif. "Alternative partnering is a key mediator of this developmental switch," says Stanton.
Stanton and Kolatkar also demonstrated that Oct4 complex formation is critical in determining the DNA motif to which it binds. A mutation in the region of Sox17 that binds Oct4 proved sufficient to shift the specificity of Oct4/Sox17 so that it binds the canonical rather than the compressed motif. Furthermore, the researchers identified similar specificity-altering mutations in Sox2, and in both cases this retargeting altered the gene activation behavior of Oct4. "Single amino acid point mutations in the Sox proteins precisely switch the developmental potential of Sox2 and Sox17," Stanton says.
These findings likely represent a broader model for gene regulation and Stanton credits his multidisciplinary team of collaborators with successfully untangling the details of this process. In future work, he hopes to explore the potential of modulating gene expression by manipulating the interactions between transcription factor pairs in a more targeted fashion.
More information: Aksoy, I., Jauch, R., Chen, J., Dyla, M., Divakar, U. et al. Oct4 switches partnering from Sox2 to Sox17 to reinterpret the enhancer code and specify endoderm. The EMBO Journal 32, 938–953 (2013). www.nature.com/emboj/journal/v … bs/emboj201331a.html
Journal information: EMBO Journal