The magic of the movies - molecules in 4D

(Phys.org) -- Computer simulations of how the body's tiniest building blocks behave are helping scientists to unlock the role of molecules in human diseases.
In a series of recent studies, researchers from Monash University's School of Biomedical Sciences have shown how important the movements and interactions of molecules are to how the body develops disease, and detects and responds to it.
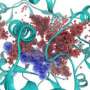
Associate Professor Ashley Buckle, who led the three projects said his team's simulations, which can take months to perform on a supercomputer, were revealing the molecular ‘choreography’ of proteins that is impossible to detect using purely experimental techniques.
"Researchers in our field have traditionally used X-ray crystallography to take a snap shot of the structure of a molecule," Associate Professor Buckle said.
"There are a number of transient interactions between molecules that we will never see using experimental techniques. These interactions explain a lot about how proteins behave and what happens when they misbehave. We need to extrapolate from the experimental data - the snapshots - and recreate the movements of molecules to explore this."
Three recent publications demonstrate the importance of this emerging research technique. Research published in PLoS Computational Biology details the key role of movement of immune system proteins in recognising infections and triggering an immune response.
"That recognition is a key event and, previously, we didn't fully understand how it happened. Simulating how the molecules move had added an extra dimension to our understanding," Associate Professor Buckle said.
A second paper, also in PLoS Computational Biology explains why an enzyme that could be an important antibiotic target works effectively when locked firmly into a group of four, but not in a “wobbly” pair. Understanding this structure could help develop treatments for an antibiotic-resistant superbug called MRSA.
A third paper, published in Biophysical Journal explains a protein mutation that causes cirrhosis of the liver.
"Using X-ray crystallography, the normal protein and the mutant protein look exactly the same. When we ran the simulation, we could see important differences in the way the molecules moved, that could explain why the mutant protein misbehaves and causes disease," Associate Professor Buckle said.
The simulations have proven so valuable that the team has set up their own supercomputer facility at Monash, known as “Orchard”, which has allowed their research to progress much faster.
"Having our own 800-core supercomputer means we can have simulations running 24/7. These simulations are so incredibly difficult that it takes months to produce a movie representing a fraction of a microsecond in real-time," Associate Professor Buckle said.
"If we were using a shared facility, the simulations that we've been running for two years would be nowhere near complete by now."
View some of the simulations at the groups' research page.
Journal information: PLoS Computational Biology , Biophysical Journal
Provided by Monash University