Mutant proteins weigh in: Researchers 'see' binding with DNA through quartz crystal microbalance
(PhysOrg.com) -- Rice University scientists have demonstrated a new way to see and quickly measure DNA/protein binding, a discovery that prompted one journal reviewer to write, "This study has made my day."
"I've never had that happen," said co-author Kathleen Matthews, Rice's Stewart Memorial Professor of Biochemistry and Cell Biology and former dean of the Wiess School of Natural Sciences.
Sibani Lisa Biswal, an assistant professor in chemical and biomolecular engineering, led the study with Matthews that made novel use of a quartz crystal microbalance (QCM) to dynamically measure the binding activity of wild-type and mutant proteins. The process let them monitor what happened in real time when DNA was first introduced to and then removed from protein molecules attached to the microbalance.
The study published this week in the American Chemical Society journal Langmuir is of interest to researchers who work in biological and chemical sensing. Co-authors include Jia Xu, a Welch postdoctoral fellow in biochemistry and cell biology, and Kai-Wei Liu, a graduate student in chemical and biomolecular engineering.
There's nothing new about QCM, a device Biswal said probably exists at most research universities. "It's mainly used for chemical and biomolecular sensing," she said. "QCM works by measuring the resonance frequency of a quartz crystal. When you apply current, it resonates, and we translate that through electronic measurements into a frequency. When you add something to the surface of the crystal, the frequency changes. That's what we're looking for."
The sensitivity is so fine that she can see the difference in a protein when a DNA molecule attaches itself to the protein. "When you apply sufficient mass to the quartz crystal, it's going to change its resonance frequency," Biswal said. "The change in mass is proportional to the change in frequency."
The researchers chose to look at an Escherichia coli protein, a lactose repressor called LacI, primarily because Matthews has the wild-type protein and multiple variants. "She has studied LacI for a long time," Biswal said. "It's a well-characterized system, and we wanted to use something we know about."
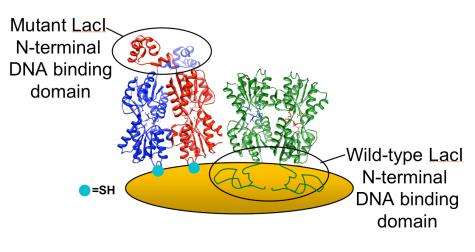
The team compared two types of LacI via QCM. The first, wild-type LacI, was used as a control because it bonds strongly to the gold-coated QCM surface using its DNA-binding domain, which spreads out like a mold and is then unable to bind DNA.
The second was a mutant form of LacI, engineered with a sulfur-based amino acid introduced far from the protein's DNA binding site. Unlike the wild-type, this mutant LacI puts down tight roots at the site of the mutation and stands like a tree, waving in the liquid breeze. "The mutation will actually orient the protein, so we can immobilize it onto a solid support," Biswal said.
Experimenting on both types, the researchers flowed liquid containing DNA and then IPTG, an inducer that releases the DNA, over the proteins bound to the QCM. In the process that took between 90 minutes and two hours, the wild-type Lacl, adsorbed by the gold surface, largely ignored the offerings.
But the mutant LacI responded with a clear change in the signal as it first grabbed passing DNA and then released it when IPTG was introduced to the stream.
"It's dynamic," Matthew said of the process. "We're flowing very small volumes of liquid over the sensor. We start with a buffer, then add the protein. When the protein binds, we can see how much goes on the surface of the crystal. Then we add the DNA and we can see that binding process with the mutant (for the wild-type, nothing happens) and then we add the IPTG and see the release."
As a bonus, the QCM technique also provides information about the viscoelastic properties of bound proteins. "It's nice that we can measure dissipation," Biswal said. "When we turn off the current, we can watch how the resonance decays -- and that tells us things like the elasticity of the material on top of the quartz."
Biswal, thinking about that reviewer, noted he had studied proteins on surfaces for the past 25 years and was delighted that one could study how mutations to a protein would change its binding properties and functionalities.
"People who develop biosensors need to be able to attach proteins onto surfaces," she said. "Our work indicates that you can't just immobilize proteins nonspecifically onto a surface. You actually do need to make some type of mutation to make sure the binding site is accessible. This platform allows us to easily screen and study what type of mutations are needed."
More information: Read the abstract at pubs.acs.org/doi/abs/10.1021/la200056h
Provided by Rice University



















