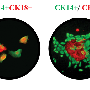
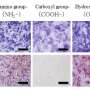
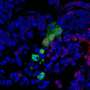
KeyGenes can predict future identity of human fetal stem cells

A snapshot of gene activity is now all that's necessary to determine what organ or tissue type that a cluster of fetal stem cells will ultimately become. An algorithm developed by a team of Dutch scientists makes it possible to match what's happening inside of an immature stem cell to known human fetal cell gene expression, thus identifying what the stem cell has the potential to be. The platform, dubbed KeyGenes and presented May 28 in Stem Cell Reports, could also prove useful for testing the quality of stem cell transplants made up of undifferentiated cells.
"One of the hardest things in stem cell biology is to determine what you have turned the cells into using a particular method of differentiation," says study senior author Susana Chuva de Sousa Lopes of the Leiden University Medical Center in The Netherlands. "We think that KeyGenes can help us develop differentiation protocols that lead to cells forming that resemble their in vivo counterparts much more closely."
CellNet, a comparable platform for mapping stem cell fate, was published by Boston researchers last summer. The difference is CellNet helps conclude the quality of procedures used to differentiate human adult tissue by comparing the tissue to microarray data (tests that see which genes are turned on or off). KeyGenes is based primarily on gene expression and integrates data from both human fetal and adult tissue to determine "identity scores" for differentiated cells.
"By collecting extensive data on gene expression during human fetal development (at different stages but also multiple organs), KeyGenes allows human stem cell derivatives to be identified and given a correct 'age' equivalent," says Leiden University co-author Christine Mummery, a board member of the International Society for Stem Cell Research.
In the clinic, the researchers believe that the tool may be helpful in cases where human development goes wrong, making it possible to identify which tissues are present in excessive amounts or are not in the right place. A platform like KeyGenes could also allow patient stem cells to be more accurately differentiated for transplant or drug testing purposes.
In the lab, KeyGenes has already led to an insight in the genes that determine a cell's fate. The genes identified are always a mix of transcription factors (molecules that determine which genes are expressed) as well as those related to cell adhesion and shape. "In addition, some of the KeyGenes identified are long non-coding RNAs, illustrating and confirming the increasingly important role recognized for this class of molecules," Lopes says.
More cell types and organs have been tested and her lab is making KeyGenes openly available to the scientific community so the platform can be used and expanded. She aims for the tool to eventually include epigenetic data sets as well to understand whether the epigenetic memory from the organ of origin remains and whether that influences the capacity of the cell to differentiate to a specific cell type.
More information: Stem Cell Reports, Roost et al.: "KeyGenes, a tool to probe tissue differentiation using a human fetal transcriptional atlas" dx.doi.org/10.1016/j.stemcr.2015.05.002
Journal information: Stem Cell Reports
Provided by Cell Press