Live bacterium depicted using X-ray laser
An international team led by Uppsala University scientists has succeeded, for the first time, in depicting intact live bacteria with an X-ray laser. This technique, now described in the journal Nature Communications, can give researchers a clearer understanding of the complex world of cells.
'If you really want to understand a cell's functions, it has to be alive,' says Professor Janos Hajdu of Uppsala University, one of the leading researchers responsible for the experiment.
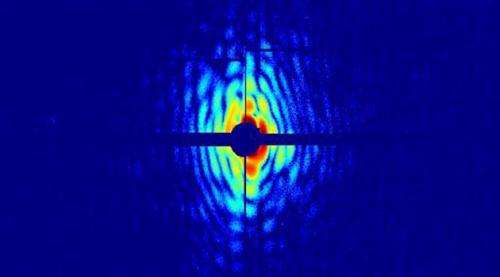
The method the researchers used in this experiment allows better resolution, in both time and space, than that obtained from the best optical microscope techniques. The new technique involves shooting a fine aerosol of cells with light pulses from an X-ray laser. This aerosol is literally a jet of living cells, thinner than a strand of hair. The ultrashort X-ray pulses spread outdiffract from individual cells, resulting in a distinct dispersion diffraction pattern that is registered by a high-speed detector. The data are then analysed with a computer programusing software developed in Uppsala, and images of the cells can be reconstructed.
X-rays destroy cells, but the ultrashort pulses and high intensity of the X-ray laser permit such rapid data collection that a correct image of the sample is obtained before it disintegrates. This technique, known as 'diffraction before destruction', has been shown previously to work with both organic and inorganic samples.
The experiment was performed with the X-ray laser of the Linac Coherent Light Source (LCLS) in the Stanford Linear Accelerator Center, California. Two species of cyanobacteria, Cyanobium gracile and Synechococcus elongatus, were used in the study. The almost cylindrical shape of these cells emerges clearly in reconstruction from diffraction data.
According to Tomas Ekeberg, a molecular biophysicist at Uppsala University and leader of the experiment, the images could have been better still if it had been possible for the detectors to handle them the data better.
'To date, we've only managed to reconstruct cells with a resolution of 76 nanometres [= millionths of a millimetre], but the data we've collected show that we can get down to 4 nanometres. That means we might be able to distinguish individual proteins, which are of that size,' Ekeberg says.
The reason for the lower resolution, Ekeberg points out, is overexposure—exactly what happens when you take photographs in excessively bright light. In future experiments, it will be possible to correct this.
'We'll be able to get much higher resolution when we're able to use a filter to reduce the overexposure,' adds Gijs van der Schot, a PhD student and principal author of the publication.
Up to now, high-resolution depiction has involved freezing cells and giving them a high radiation dose that kills them during the data collection. The cells have also often been coated with a thin layer of metal to improve the contrast. Static images of cells also normally require long exposure times.
The research team's new method can show the structure of living cells, virtually instantaneously. Every image takes only a few femtoseconds (one femtosecond being a millionth billionth of a second). This kind of tool could help scientists gain a better grasp of hitherto unknown details of cell function and behaviour.
'Processes like cell division and protein folding could be observed in real time. The technique also makes possible future 3D modelling of processes in the cell, which could give us important insights into complicated disease progression,' Ekeberg explains.
The team is now planning to fine-tune the depiction method with further experiments, and hope to achieve images with a considerably higher resolution.
'There's a huge difference between images produced by means of this technique and those from traditional optical microscopy of living cells,' says Janos Hajdu.
'Few people thought this was feasible.'
Publication details
"Imaging single cells in a beam of live cyanobacteria with an X-ray laser." Nature Communications 6, Article number: 5704 DOI: 10.1038/ncomms6704
Journal information: Nature Communications
Provided by Uppsala University



















