Scientists "resurrect" ancient proteins to learn about primordial life on Earth

Geological evidence tells us that ancient Earth probably looked and felt very different from the planet we all recognize today. Billions of years ago, our world was a comparatively harsh place. Earth likely had a hotter climate, acidic oceans and an atmosphere loaded with carbon dioxide. The fact that manmade climate change, through carbon dioxide pollution, is re-introducing such hotter, acidified conditions demonstrates their intertwinement.
More recently, the life sciences have begun buttressing these notions of primordial Earth. Thanks to advances in a niche field called paleobiochemistry, researchers in the last decade have started to "resurrect" ancient proteins. Studying these proteins' properties is offering us glimpses of what life was like in bygone epochs.
The results so far are compelling. Take, for example, beta lactamase proteins, which first evolved between 2 to 3 billion years ago. These ancient proteins actually remain more stable and work better in hot spring-like temperatures of between 130 and 150 degrees Fahrenheit (54 and 66 degrees Celsius) compared to their modern counterparts. Other proteins, called thioredoxins, originated 4 billion years ago at the time of life's origin, and these ancient proteins stay active in acidities that would break down many modern proteins. Findings of this sort help paint a portrait of life prior to 500 million years ago in the vast era known as the Precambrian.
"Molecular resurrection studies provide a new line of evidence supporting geological models that suggest that the Precambrian Earth hosted a hotter and more acidic ocean than its modern counterpart," Eric Gaucher, a pioneering paleobiochemist and a professor biology at Georgia Tech. "Early life was adapted to this environment."
Paleobiochemistry should have much more to eventually say on this topic. Toward this end, Gaucher and colleagues at the University of Granada in Spain have a new paper in the June 2014 issue of the scientific journal Proteins: Structure, Function, and Bioinformatics. The study compares two common techniques used in paleobiochemistry that have potential biotechnology applications, such as finding ways of dealing with the scourge of antibiotic resistance. The two methods allow scientists to extrapolate the composition of proteins from eons ago.
Deciphering the development of biota on Earth is important not only for piecing together our planet's past—and thus its potential future—but also for gauging where else life might arise in the cosmos.
"Knowing how life originated and diversified on early Earth provides us with a perspective on the conditions that support primitive life," said Gaucher. "This information can better inform our decisions to search for life on other planets."
Breathing new life into old proteins
Living creatures use proteins for much of life's business. These molecules form many of the structural components of cells and facilitate the chemistry for powering them. Made from combinations of any of 20 amino acid building blocks, proteins come in an almost endless variety of complexity and function.
"It is remarkable to think that there are billions of different proteins contained within all organisms on modern Earth," said Gaucher. "Yet, these proteins are composed of the same building blocks, only arranged in different configurations or sequences."
Researchers have compiled huge databases full of the proteins' amino acid sequences. The sequences have changed over evolutionary history. By comparing today's sequences to each other, scientists can get a good idea of the sequence of an ancestral protein from which the modern versions descended.
The concept is rather like that of tracing modern languages back to older, source languages, as Gaucher has previously explained to Astrobiology Magazine. By comparing several European languages, for instance, one would discern that French, Italian, Portuguese, Romanian and Spanish all have clear Latin roots.
"Molecular resurrection studies use a top-down method, whereby modern biological information is used to infer ancient biology," said Gaucher. "Studying this ancient biology gets us closer to the origins of life itself."
A consensus approach
One method is called consensus-sequence engineering, and is the more simplistic of the two. Scientists just plug the sequence of a protein of interest into a protein database. The query returns a large number of "hits," or analogous sequences.
"These sequences likely correspond to modern proteins that are evolutionarily related to the query protein," said Valeria Risso, lead author of the new Proteins paper and a chemist at the University of Granada.
From there, Risso and colleagues gather statistics on the particular amino acids that appear at corresponding positions on the analogous proteins. Whichever amino acid pops up most frequently is deemed the "consensus" amino acid.
In theory, this consensus amino acid had previously occurred at the sequence location earlier on in evolutionary history, before mutations led to divergent, modern sequences. This makes sense, because nature is conservative. Evolution should favor keeping a sequence that works versus a mutated one that doesn't. Every now and then, of course, a mutation will "earn its keep" by providing the organism with an advantage or—at the very least—not hinder its possessor from reproducing.
The consensus-seeking task is completed for every amino acid position. The next step: artificially generating the consensus protein in the lab. This is done by introducing a modified gene into a model organism, such as the bacterium E. coli. The organisms handily cranks out the protein through natural means. The new, yet old-fangled protein can then be put through its paces by seeing how it chemically reacts in certain conditions.
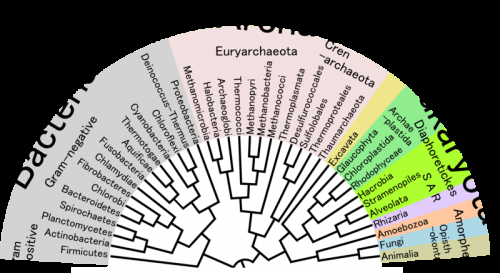
Tree of life
The second method more closely follows the historical linguist analogy. It involves creating what is known as a phylogenetic tree. Essentially, the protein sequences are compared, as before, but they go through an evolutionary model-based analysis to search for "nodes," or branching points.
The nodes' sequences represent the last common ancestor for the species that subsequently split off, on down to modernity. Another way to think of this phylogenetic tree method is that it is essentially a genealogy.
The node sequences are inferred and compiled. The proteins encoded by the reconstructed sequences are then synthesized in the laboratory—"laboratory resurrection," as it's called. The protein, as with consensus-sequence engineering, is produced by a model organism and its properties are then assessed.
Hopefully, the resulting protein's properties, when compared to its descendant proteins, should fit like a link in a logical evolutionary chain.
"These properties are expected to 'tell' a story that makes sense in biological terms by providing a convincing evolutionary narrative," said Gaucher.
An example of this proteinaceous storytelling: the fact that ancient proteins, as mentioned before, seem optimized for the high-heat, high-acidity environmental conditions which geology suggests characterized the young Earth.
Both paleobiochemistry techniques seek to restore proteins long lost through the vicissitudes of evolution. But is the consensus technique as good at recovering "real" primordial proteins as the phylogenetic approach? The new paper aimed to answer this question.
Dueling time machines
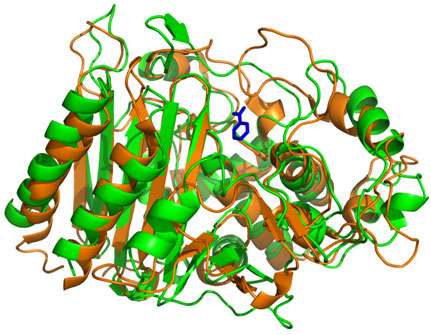
The researchers compared properties of a beta lactamase protein yielded from consensus sequence and phylogenetic sequence methods. Beta lactamase is a primary means of antibiotic resistance. It allows an organism to persevere against the lactam class of antibiotics; we rely on numerous lactam drugs, such as penicillin, to fight off infections.
Three consensus variants were created for the study. Sequence-wise, they were indeed quite like the sequences made by the more rigorous, phylogenetic approach.
However, the consensus-sequence derived proteins were not as stable as the phylogenetic proteins. Nor did they partner up with as many other relevant molecules. This is a trending trait of ancient proteins, which according to theory, started out as generalists, then honed and specialized over the course of evolution. Though the consensus sequence proteins differed by just a few amino acids, important differences in functionality followed.
Overall, consensus engineering does not look like the best way to work backwards toward discovering how ancient life worked, either from a biotechnology or an astrobiology standpoint.
"Consensus certainly remains an interesting approach in protein engineering," said paper-coauthor Jose M. Sanchez-Ruiz, also a chemist at the University of Granada. Nevertheless, Sanchez-Ruiz added, the study's "results support ancestral reconstruction and resurrection as a more efficient procedure to obtain proteins with extreme and useful properties."
Life, decoded
Learning more about primordial life will open up a lot of avenues for science. On a fundamental level, reconstructing life back through the ages gets us more familiar with the parts and pieces biology requires.
"Analogous to the engineering adage that you cannot understand something unless you can build it, a fuller understanding of life will only come when we can build life," said Gaucher.

Gauging what sorts of ingredients and environments were conducive to life forming on Earth will inform astrobiological ambitions. Knowing what to look for on future missions to potential places for life, like Jupiter's moon Europa, will be one benefit of a more complete picture of early Earth's microbes.
Better yet than toying with individual proteins, though, would be sizing up a whole organism. And stay tuned: by building on their success with phylogenetics, Gaucher and colleagues hope to be able to bring ancient bacteria and archaea back from the dead.
"Although the majority of resurrection studies currently focus on resurrecting one or two protein families at a time," Gaucher said, "we anticipate that we will be able to resurrect a complete ancestral genome in the near future and jump-start this genome using modern life to, in essence, resurrect long extinct forms of life."
More information: Risso, V. A., Gavira, J. A., Gaucher, E. A. and Sanchez-Ruiz, J. M. (2014), "Phenotypic comparisons of consensus variants versus laboratory resurrections of Precambrian proteins." Proteins, 82: 887–896. doi: 10.1002/prot.24575
Source: Astrobio.net
This story is republished courtesy of NASA's Astrobiology Magazine. Explore the Earth and beyond at www.astrobio.net .











.jpg)






