Knowing what to keep and what to trash: How an enzyme distinguishes cellular messages
Every once in a while, we are forced to sort that stack of papers on the kitchen counter. Interspersed between the expired coupons and dozens of takeout menus are important documents like your car insurance or electric bill. So it isn't an option to simply drop it all in the trash at once – you need to read through the messages to be sure that you don't lose vital information.
In the cell, proteins similarly read through messages to distinguish what needs to be saved and what needs to be discarded. But, here, the process takes on a much more important role. More than just clutter, messages that are marked for disposal can drastically alter the fate of a cell. In fact, stem cells use just such a mechanism to maintain their identity. So how does a protein detect the difference between two seemingly similar messages? Today, a team of Cold Spring Harbor Laboratory (CSHL) scientists, led by Professor and Howard Hughes Medical Institute Investigator Leemor Joshua-Tor, describe how the protein Dis3l2 uses numerous recognition sites to capture messages that are flagged for decay.
Dis3l2 is a molecular machine that helps to preserve the character of stem cells. It serves as the executioner of an elegant pathway that prevents stem cells from changing into other cell types. The protein does this by acting like a garbage disposal for messages in the cell, cutting them up until they no longer encode useful information. But Dis3l2 is necessarily highly specific. While it must degrade messages that would alter the fate of the stem cell, discarding the wrong message could have devastating consequences.
Therefore, Dis3l2 only targets specific messages that have been marked with a molecular flag, known as a "poly-U" chain. The enzyme ignores the majority of messages in the cell – those that go on to encode proteins and other critical messages – whose ends are decorated with a different type of chain, called "poly-A" tail.
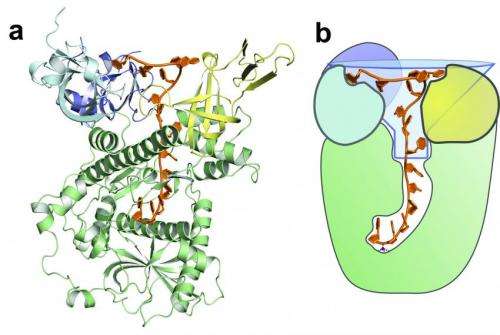
The CSHL scientists set out to understand how Dis3l2 is able to read and distinguish between these two chains. In work published today in Nature, they used a type of molecular photography, known as X-ray crystallography, to observe the structure of Dis3l2 while bound to a poly-U chain. "We saw that the enzyme looks a lot like funnel, quite wide at the top and narrow at the base," says Joshua-Tor. "The poly-U chain inserts itself into the depths of this funnel while the rest of the bulky message can remain in the wide mouth at the top."
But how does the enzyme "read" the poly-U chain? Christopher Faehnle, PhD and Jack Walleshauser, lead authors on the paper, found that the interior of the funnel contains more than a dozen contacts that interact specifically with the poly-U chain. "Together, all of these points create a sticky web that holds the poly-U sequence deep within the enzyme," says Faehnle. "But other chains don't interact – they can slide right out. It has helped us understand how an enzyme can differentiate between two sequences in the cell."
More than that, the work provides insight into how a stem cell maintains its identity. "Misregulation of any step in this pathway leads to developmental disorders and cancer," says Joshua-Tor. "We now have a much better appreciation of the terminal step, a critical point of control."
More information: "Mechanism of Dis3l2 substrate recognition in the Lin28–let-7 pathway" appears online in Nature on August 3, 2014. The authors are: Christopher Faehnle, Jack Walleshauser and Leemor Joshua-Tor. The paper can be obtained online: dx.doi.org/10.1038/nature13553
Journal information: Nature
Provided by Cold Spring Harbor Laboratory