A particle dynamics model may provide insight into diseases and deformities linked to disruptions in cell development
A computer-based model of cell particle dynamics shows that the lopsided torque produced by forces within a cell may explain the previously puzzling motion and shape of rotating cell pairs. The model provides novel insights that challenge current thinking about the causes of developmental abnormalities and of cancer and other diseases linked to disrupted cell rotation.
A crucial feature of the model of key particle motions, developed by Fong Yew Leong at the A*STAR Institute of High Performance Computing in Singapore, is that it spontaneously produces cells with the same shape and rotational characteristics as real cells.
During the development of multicellular organisms, cells rotate around each other in a coordinated manner. This rotation does not occur in some cancer cells, which implies that disrupting normal cell movement may influence disease as well as development.
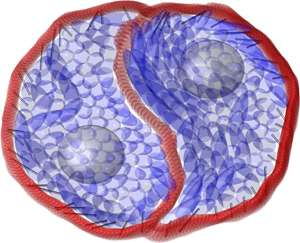
Isolated cells growing on flat surfaces tend not to rotate. However, two joined cells can rotate spontaneously and continuously, and will often develop a sigmoidal or 'S-shaped' interface that resembles the yin and yang symbol (see image and video). As the cells rotate, they appear to 'moonwalk' around one another; each cell moves in the opposite direction to its protrusion into the other cell and in the reverse manner to cells moving on their own.
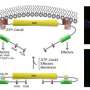
Leong modeled the cells in two dimensions as an assembly of cytoplasm particles surrounded by a cell membrane. As cell movement is driven by the interaction of actin and myosin protein filaments in the cell's cytoskeleton, Leong designed the model to include the formation and degradation of actin and myosin chains attached to the inner cell membrane.
The simulation showed the interaction of actin and myosin within the cell—known as 'undirected actomyosin forcing'—which is powerful enough to generate the shape and movement of the cells. Crucially, as Leong explains, "forces that are angled toward the cell membrane lead to an unbalanced torque that rotates the cell." He also considers the spontaneous emergence of the torque, due to the tilted forces, to be the most significant insight provided by his model.
"The next step is to develop a three-dimensional model that explains cell cluster rotation in vivo, rather than just on a two-dimensional surface," says Leong. He hopes future iterations of the rather simple current model will help explain the real-life complexity of the movements of multiple cells and ultimately advance approaches to addressing developmental abnormalities and diseases linked to the disruption of cell movement.
More information: Leong, F. Y. "Physical explanation of coupled cell-cell rotational behavior and interfacial morphology: A particle dynamics model." Biophysical Journal 105, 2301–2311 (2013). DOI: 10.1016/j.bpj.2013.09.051
Journal information: Biophysical Journal