Tiny algae shed light on photosynthesis as a dynamic property
Many of the world's most important photosynthetic eukaryotes such as plants got their light-harnessing organelles (chloroplasts) indirectly from other organisms through endosymbiosis. In some instances, this resulted in algae with multiple, distinct genomes, some in residual organelles (nucleomorphs). To better understand why nucleomorphs persist after endosymbiosis, an international team including researchers at the DOE Joint Genome Institute collaborated to sequence and analyze two tiny algae. Their report appeared online Nov. 29, 2012 in Nature.
One of the first chemical reactions children learn is the recipe for photosynthesis, combining carbon dioxide, water and solar energy to produce organic compounds. Many of the world's most important photosynthetic eukaryotes such as plants did not develop the ability to combine these ingredients themselves. Rather, they got their light-harnessing organelles—chloroplasts—indirectly by stealing them from other organisms. In some instances, this has resulted in algae with multiple, distinct genomes, the evolutionary equivalent of a "turducken."
Chloroplasts originally evolved from photosynthetic bacteria by primary endosymbiosis, in which a bacterium or other prokaryote is engulfed by a eukaryotic host. The chloroplasts of red and green algae have subsequently come to reside within other, previously non-photosynthetic eukaryotes by secondary endosymbiosis. Such events have contributed to the global diversity of photosynthetic organisms that play a crucial role in regulating and maintaining the global carbon cycle. In most organisms that acquired photosynthesis by this mechanism, the nucleus from the ingested algal cell has disappeared, but in some cases it persists as a residual organelle known as a nucleomorph. Such organisms have four distinct genomes.
To better understand the process of secondary endosymbiosis and why nucleomorphs persist in some organisms, an international team composed of 73 researchers at 27 institutions, including the U.S. Department of Energy Joint Genome Institute (DOE JGI), collaborated to sequence and analyze the genomes and transcriptomes (the expressed genes) of two tiny algae. The team led by John Archibald of Canada's Dalhousie University published their findings on the algae Bigelowellia natans and Guillardia theta online November 29, 2012 in Nature.
Archibald compared these algae to Russian nesting dolls with "sophisticated sub-cellular protein-targeting machinery" and four genomes derived from the two eukaryotes that merged over time. Approximately 50 percent of the genes in both genomes are 'unique' with no obvious counterpart in other organisms," he added. "This indicates just how different they are from characterized species."
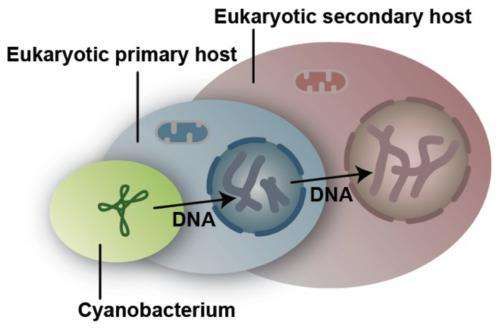
DOE JGI Fungal Genomics Program head Igor Grigoriev called B. natans and G. theta "living fossils" because of the remnant nucleomorph. He added that algae are relevant to the Department of Energy Office of Science research portfolio for their potential applications in the fields of bioenergy and environment, noting that the DOE JGI has published over 75 percent of the publicly available algal genomes. "Iterations of endosymbiosis have led to a global diversity of these primary producers. Sequencing these two algae, the first cryptophyte and the first chlorarachniophyte sequenced, helped us to fill in the gaps in the Eukaryotic Tree of Life, and obtain additional references for better understanding of eukaryotic evolution."
Archibald said that "G. theta and B. natans both possess a surprisingly complex suite of enzymes involved in carbon metabolism, and thus represent a useful resource for scientists engaged in both basic and applied research, including photosynthesis, sub-cellular trafficking and biofuels development."
The DOE JGI sequenced the genomes of B. natans (95 million nucleotides or bases: Mb) and G. theta (87 Mb) from single cell isolates provided by Bigelow Laboratory for Ocean Sciences as part of the 2007 Community Sequencing Program portfolio. In addition, the transcriptomes were sequenced separately by the National Center for Genome Resources in New Mexico. Among the team's findings is an answer to the question of why nucleomorphs still exist.
"The reason for the persistence of nucleomorphs in both organisms appears to be surprisingly simple: they are no longer able to transfer their DNA to the host cell nucleus by the process of endosymbiotic gene transfer," said Archibald. Unlike most other secondarily photosynthetic eukaryotes in which the endosymbiont's genetic matter has completely migrated over to the host, in cryptophytes and chlorarachniophytes the nucleus and chloroplast from the engulfed algae remain partitioned off from the host cell. "As a consequence," he added," genetic and biochemical mosaicism is rampant in G. theta and B. natans."
Both researchers highlighted the unexpected finding of alternative splicing in B. natans. Grigoriev noted that the phenomenon is one typical for higher eukaryotes, and Archibald added that the levels "greatly exceed that seen in the model plant Arabidopsis and on par with the human cerebral cortex, unprecedented and truly remarkable for a unicellular organism. This challenges the paradigm that complex alternative splicing is a phenomenon limited to sophisticated multicellular organisms."
"The evolution of chloroplasts, the photosynthetic compartments of plants and algal cells, is complex but has had a profound effect on our planet," said Chris Howe, Professor of Plant and Microbial Biochemistry at Cambridge University in England. "This paper gives us fascinating insights into how host and nucleomorph genomes have been remodeled during evolution. As well as providing a goldmine of information on the general biology of these organisms, the paper shows us that the nucleomorph genomes have probably persisted simply because the mechanism for transfer of genes to the nucleus was closed off, rather than because nucleomorphs had to be retained as separate entities. The paper also shows us that the evolutionary history of the nucleomorph-containing organisms was even more complex than we thought, with evidence for genes from many different sources in their nuclei. Overall, it has provided important insights into the fundamental processes of cell symbiosis and genome reshaping that have produced some of the most important organisms we see today."
Journal information: Nature
Provided by DOE/Joint Genome Institute















