Researchers identify structure that allows bacteria to resist drugs

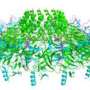
A research team led by Edward Yu of Iowa State University and the Ames Laboratory has discovered the crystal structures of pumps that remove heavy metal toxins from bacteria, making them resistant to antibiotics.
The findings are published in the Sept. 23 issue of the journal Nature.
Yu - an Iowa State associate professor of chemistry, of physics and astronomy, of biochemistry, biophysics and molecular biology and an associate of the U.S. Department of Energy's Ames Laboratory - said the finding gives researchers a better understanding of bacterial resistance to antibiotics. Ultimately it could help drug researchers develop treatments to combat that resistance.
To make their findings, the researchers purified and crystallized the membrane proteins that make up an efflux pump of E. coli bacteria. The researchers prepared some samples that contained the toxic heavy metals copper and silver and some that did not.
The researchers used X-ray crystallography to compare the various structures, identify the differences and understand the mechanism that removes heavy-metal toxins from cells.
Their paper specifically describes the crystal structure of CusA, one of three parts of the pumps responsible for removing toxins from bacteria. Yu said CusA is an inner membrane transporter which belongs to the resistance-nodulation-division protein superfamily. It consists of 1,047 amino acid residues and spans the inner membrane 12 times.
What those pumps do, Yu wrote in a summary of his research, is "recognize and actively export these substances out of bacterial cells, thereby allowing the bugs to survive in extremely toxic conditions."
The research project was supported by the National Institutes of Health. In addition to Yu, the research team includes Robert Jernigan, an Iowa State professor of biochemistry, biophysics and molecular biology and director of Iowa State's Laurence H. Baker Center for
Bioinformatics and Biological Statistics; Kanagalaghatta Rajashankar, the operations team leader for the Northeastern Collaborative Access Team facility at Argonne National Laboratory in Argonne, Ill., that's managed by Cornell University in Ithaca, N.Y.; Iowa State post-doctoral researchers Feng Long and Chih-Chia Su; and Iowa State graduate students Michael Zimmermann and Scott Boyken.
"This work reports the first detailed structure of a unique heavy metal transporter that enables bacteria to survive the toxic effects of silver and copper," said Jean Chin, Ph.D., who oversees this and other structural biology grants at the National Institutes of Health. "By detailing the exact steps that a metal ion is likely to take through the transporter, this study suggests how we might block the pathway and render pathogenic bacteria sensitive to heavy metal toxins."
Yu, who has been studying bacterial resistance to antibiotics for nearly a decade, said direct information about how bacteria handle heavy-metal toxins is important information for biomedical researchers.
"We want to understand the mechanisms of these heavy-metal pumps," he said. "And that could allow biotechnology researchers to make inhibitors to stop the pump and the antibiotic resistance."
Provided by Iowa State University