Breakthrough in tracking a cause of chronic infections
(PhysOrg.com) -- Northeastern University researchers have identified a protein that enables bacterial cells to survive antibiotic treatment.
This discovery by biology professor Kim Lewis and his team, published in the February 22 issue of the journal PloS Biology, reveals for the first time the mechanism by which bacteria maintain a chronic infection in spite of aggressive therapy with antibiotics.
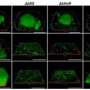
Lewis and his team previously discovered that pathogens responsible for chronic infections form small populations of dormant cells, called persisters, which are not killed by antibiotics. Antibiotics function only on active cells, so when treatment ceases, persisters grow and repopulate, causing a relapse. The protein identified by the Northeastern researchers, known as TisB, triggers a cellular response that leads to the development of persister cells.
“The paradox of chronic infections is that, in many cases, pathogens aren’t resistant to antibiotics, yet the antibiotics don’t work,” said Lewis, director of Northeastern’s Antimicrobial Discovery Center. “We now better understand what happens inside the cell that enables pathogens to become inactive and cause relapsing infections.”
The research was conducted using E. coli, a common bacterium found in food and water supplies, and a frequently prescribed antibiotic, ciprofloxacin. Lewis and his team discovered that while the antibiotic was killing most bacteria, it also induced production of TisB in some bacterial cells, which resulted in the formation of persisters.
These findings represent the first time scientists have been able to identify — at the molecular level — how persister cells are formed, said Lewis.
“This is the first step towards understanding the complex workings of how cells evolve to protect themselves,” said Lewis. “The more we know, the better equipped we will be to develop counter measures and effective treatments against bacterial infections.”
Provided by Northeastern University