This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Probing small molecule-RNA interactions by looking through the FOREST
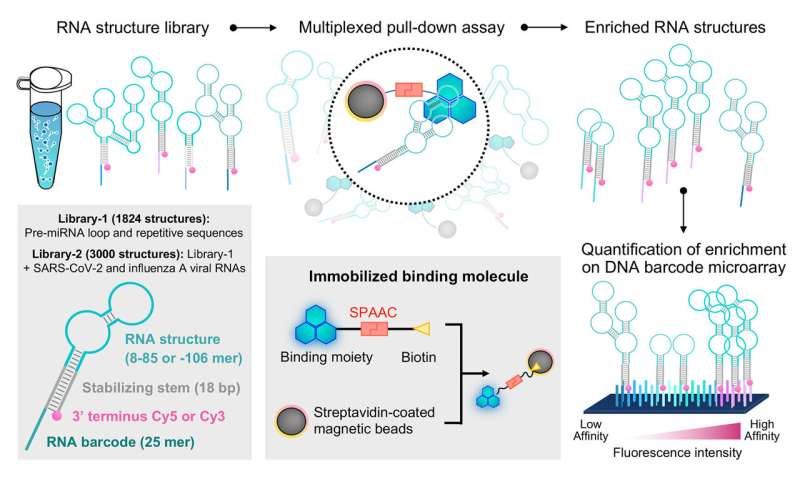
A team of researchers has recently demonstrated the utility of employing a previously established screening system to probe the interactions between small molecules and RNA. Their study is published in Communications Chemistry.
The team was led by Professor Hirohide Saito (Department of Life Science Frontiers, CiRA), Dr. Kaoru R. Komatsu (a former Ph.D. student in CiRA), Associate Professor Kazumitsu Onizuka, and Professor Fumi Nagatsugi (Institute of Multidisciplinary Research for Advanced Materials, Tohoku University).
From recent SARS-CoV-2 mRNA vaccines to combat the COVID-19 pandemic to risdiplam, an RNA splicing modifier approved by the US Food and Drug Administration for spinal muscular atrophy, the word "RNA" has entered into common language as it represents both new classes of therapeutic agents and drug targets.
However, our understanding of how various RNA sequences and, in turn, structures dictate interactions with small molecules such as drug compounds or large biomolecules such as proteins remains incomplete.
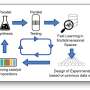
In a previous study, Professor Saito and his research team designed a system called folded RNA element profiling with structure library, or FOREST, to examine the molecular details of how RNA interacts with known RNA-binding proteins.
For this new study, through a collaboration with researchers from Tohoku University, the joint research team illustrated how FOREST can be utilized to analyze small molecular interactions with RNA.
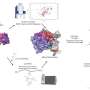
The research team first validated the applicability of the FOREST approach to small molecules by examining how an RNA structure library interacts with known small molecular RNA interactors: G-clamp and thiazine orange (TO) derivatives.
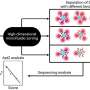
The researchers isolated RNA structures bound to G-clamp from a library comprising more than 1,800 RNA sequences derived from human pre-miRNAs and other repetitive and control sequences. Because these RNA structures are linked to a fluorescent dye and RNA barcode, they could be readily decoded and quantified by a DNA microarray with sequences complementary to the RNA barcodes to provide a quantitative analysis of how RNA structures interact with the specific small molecule of interest.
As expected, they identified preferential binding to guanosine (G)-containing single and double-stranded RNA sequences (ssRNA and dsRNA, respectively) by G-clamp. From the RNA structure library, the research team chose sequences showing high-, intermediate-, or low-affinity binding to G-clamp for validation by an independent fluorescence-based experiment that directly measures apparent dissociation constants of individual interactions.
Remarkably, they observed a good correlation between the relative binding affinity estimated by the FOREST approach and the apparent dissociation constants determined by the fluorescence-based binding assay, indicating the high robustness of this method for quantifying small molecule-RNA interactions.
Furthermore, by mutating a specific RNA loop structure with multiple guanosines at different locations, they discovered that G-clamp does not interact with all guanosines on the loop equally but that additional structural context can influence the interaction.
Conversely, TO derivatives are commonly used probes for fluorescent indicator displacement (FID) assays. The researchers next mixed TO and TO-3 separately with an expanded RNA structure library containing additional sequences derived from SARS-CoV-2 and influenza A virus RNAs to better characterize TO derivatives for RNA measurements.
As expected, whereas there was no correlation between binding profiles of RNA structures interacting with G-clamp and TO derivatives, TO and TO-3 shared similar binding profiles with some minor distinctions.
Further comparisons between TO-N3, TO-N3-2, and TO-3-N3 revealed that linker position has a modest influence over RNA binding profiles. In addition, based on these binding profiles, the researchers discerned some base and loop positional preferences that TO derivatives have when interacting with RNA structures.
The research team additionally extended their analysis of TO derivatives by comparing relative binding affinities determined by FOREST to the apparent dissociation constants measured for commercially available fluorescent nucleic indicators, TO-PRO-1 and TO-PRO-3, by the fluorescence-based binding assay.
Through this analysis, they revealed that while TO-N3-2 may more accurately portray the binding profile of TO-PRO1 compared to TO-3-N3, both TO-N3-2 and TO-3-N3 simulate TO-PRO-3 about equally well, thus providing crucial structural insights for improving pairings of target RNA and fluorescent indicators for FID assays.
Using the binding profiles determined for TO derivatives, the research team selected combinations of fluorescent indicators (TO-PRO-1 or TO-PRO-3) and pre-miRNA sequences previously shown to be dysregulated in tumors with intermediate binding affinities for FID assays.
As a proof-of-concept, the researchers screened a commercially available chemical library with 118 compounds to identify small molecules capable of interacting with disease-associated pre-miRNAs. Through this effort, they identified baicalein (Bai), myricetin (Myr), chelerythrine chloride (Che), and AS 602801 (AS) as candidate hit compounds. Whereas Myr and Che are known to bind DNA and RNA, this was the first demonstration of AS as a nucleic acid interactor.
Notably, the researchers observed different results when TO-PRO-1 or TO-PRO-3 was used as the fluorescent indicator, thus suggesting that distinct indicators should be employed to avoid false positive and negative identifications. Further examination of AS confirmed binding to several human pre-miRNAs of interest, but the researchers also noticed the compound to exhibit strong light-up properties when interacting with RNA.
Structural examination of the compound suggests it contains a chemical moiety likely responsible for the light-up properties, making it a compound of interest for further development into a novel RNA interactor and fluorescent probe.
In this study, the joint research effort once again illustrates the applicability of the FOREST methodology, not only for inspecting RNA-protein interactions but also for investigating the fine details of interactions between RNA and small molecules.
Given the vast potential for RNA as a new therapeutic approach in next-generation medicine, the ability to systemically characterize small molecule-RNA interactions on large scales will have enormous impacts on basic RNA research and the translation of that knowledge into therapies.
More information: Ryosuke Nagasawa et al, Large-scale analysis of small molecule-RNA interactions using multiplexed RNA structure libraries, Communications Chemistry (2024). DOI: 10.1038/s42004-024-01181-8
Journal information: Communications Chemistry
Provided by Kyoto University