This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
International study produces a comprehensive 'tree of life' for flowering plants
With their own botanical collection material and their research knowledge on the evolution of cruciferous plants (plants of the cabbage family), bioscientists at Heidelberg University have contributed to a large-scale international study that has produced a comprehensive "tree of life" for flowering plants.
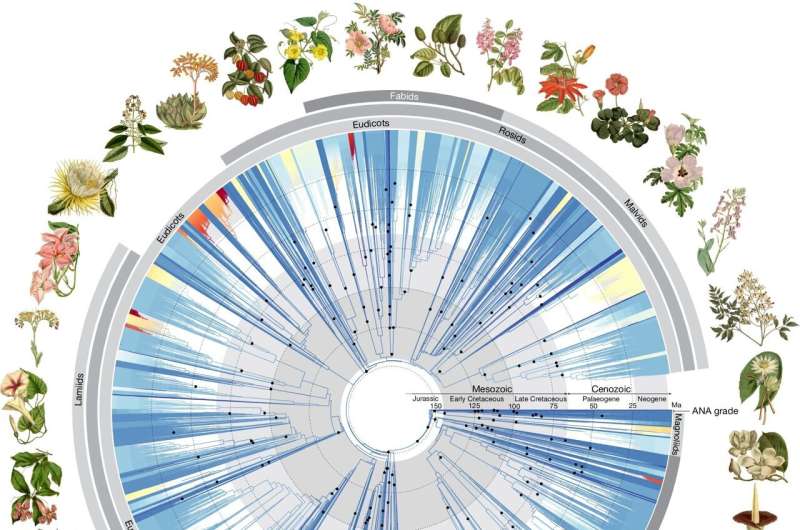
For this purpose, researchers worldwide—led by the Royal Botanic Gardens in Kew (United Kingdom)—analyzed the genetic information of more than 9,500 species from almost 8,000 genera. Besides well-known plant types found on Earth today they also examined the genetic codes of centuries-old specimens and already extinct examples.
For their participation in the "Tree of Life" project, the Heidelberg scientists from the Centre for Organismal Studies were able to use comprehensive research material from living collections, seed collections, and the herbarium.
At the Centre for Organismal Studies (COS) of Heidelberg University, the Biodiversity and Plant Systematics Department, headed by Prof. Dr. Marcus Koch, does research on the origin of species and biological diversity, as well as elucidating and describing the underlying evolutionary processes.
Plants of the cabbage family are a major focus. Besides cultivated plants, they include scientific model plants such as thale cress, also known as Arabidopsis thaliana. For their research studies the Heidelberg scientists draw on plant material with a documented history and origin.
"In the past over 25 years, we have collated this material on many voyages of discovery and expeditions and have deposited it in our curated collections," says Prof. Koch, who is also director of Heidelberg University's Botanic Garden, which comprises almost 10,000 species in living cultivation.
Particularly interesting to Marcus Koch and his team are herbaria, in which plants and plant parts are conserved for scientific purposes in dried or pressed form. The herbarium in Heidelberg contains almost 500,000 specimens. "Even centuries later the DNA, the genetic information, of dried plants can be isolated and used for evolutionary analyzes," explains Prof. Koch.
Also crucial to the research work are seed banks. Under optimum circumstances, even centuries-old plant material can be made to germinate again, says the scientist, who has engaged in teaching and research at Heidelberg University since 2003 as Professor for Plant Systematics, Biodiversity and Evolution.
Regarding the cabbage family, the researchers at the COS have built up not only a comprehensive collection of herbarium specimens and seeds with thousands of samples but also an extensive scientific database called BrassiBase. In addition to their own research material, they use stocks available in other German and international collections.
The Heidelberg research knowledge on the evolutionary processes of the cruciferous plants and the origin of their species flowed into the "tree of life" for flowering plants, which has just appeared. Flowering plants constitute around 90 percent of all known plants on land, are to be found practically everywhere on Earth, and are used as food, raw material, or a source of energy.
Having emerged over 140 million years ago, the question of how they were able to develop this "dominance" vis-à-vis other plants has occupied researchers to this day. The "tree of life"—among the 9,500 species analyzed were 800 flowering plants alone whose DNA had so far not yet been sequenced—now allows us to have new insights into their origin and relationships.
The project initiators underline that the data will contribute to identifying new species, refining plant classification, uncovering new medicinal compounds, and conserving plants in the face of climate change and biodiversity loss. 279 researchers from 138 organizations worldwide collaborated in the large-scale study.
The paper is published in the journal Nature.
More information: William Baker, Phylogenomics and the rise of the angiosperms, Nature (2024). DOI: 10.1038/s41586-024-07324-0. www.nature.com/articles/s41586-024-07324-0
Journal information: Nature
Provided by Heidelberg University