Traveling salesman uncorks synthetic biology bottleneck
Researchers have created a computer program that will open a challenging field in synthetic biology to the entire world.
In the past decade, billions of dollars have been spent on technology that can quickly and inexpensively read and write DNA to synthesize and manipulate polypeptides and proteins.
That technology, however, stumbles when it encounters a repetitive genetic recipe. This includes many natural and synthetic materials used for a range of applications from biological adhesives to synthetic silk. Like someone struggling with an "impossible" jigsaw puzzle, synthesizers have trouble determining which genetic piece goes where when many of the building blocks look the same.
Scientists from Duke University have removed this hurdle by developing a freely available computer program based on the "traveling salesman" mathematics problem. Synthetic biologists can now find the least-repetitive genetic code to build the molecule they want to study. The researchers say their program will allow those with limited resources or expertise to easily explore synthetic biomaterials that were once available to only a small fraction of the field.
The results appear in Nature Materials, January 4, 2016.
"Synthesizing and working with highly repetitive polypeptides is a very challenging and tedious process, which has long been a barrier to entering the field," said Ashutosh Chilkoti, the Theo Pilkington Professor of Biomedical Engineering and chair of the biomedical engineering department at Duke. "But with the help of our new tool, what used to take researchers months of work can now be ordered online by anyone for about $100 and the genes received in a few weeks, making repetitive polypeptides much easier to study."
Every protein and polypeptide is based on the sequencing of two or more amino acids. The genetic recipe for an individual amino acid—called a codon—is three letters of DNA long. But nature has 61 codons that produce 20 amino acids, meaning there are multiple codons that yield a given amino acid.
Because synthetic biologists can get the same amino acid from multiple codons, they can avoid troublesome DNA repeats by swapping in different codons that achieve the same effect. The challenge is finding the least repetitive genetic code that still makes the desired polypeptide or protein.
"I always thought there was a potential solution, that there must be a way of mathematically figuring it out," said Chilkoti. "I had offered this problem to graduate students before, but nobody wanted to tackle it because it requires a particular combination of high-level math, computer science and molecular biology. But Nicholas Tang was the right guy."
After studying the problem in detail, Nicholas Tang, a doctoral candidate in Chilkoti's laboratory, discovered that the solution is a version of the "traveling salesman" mathematics problem. The classic question is, given a map with a set of cities to visit, what is the shortest route possible that hits every city exactly once before returning to the original city?
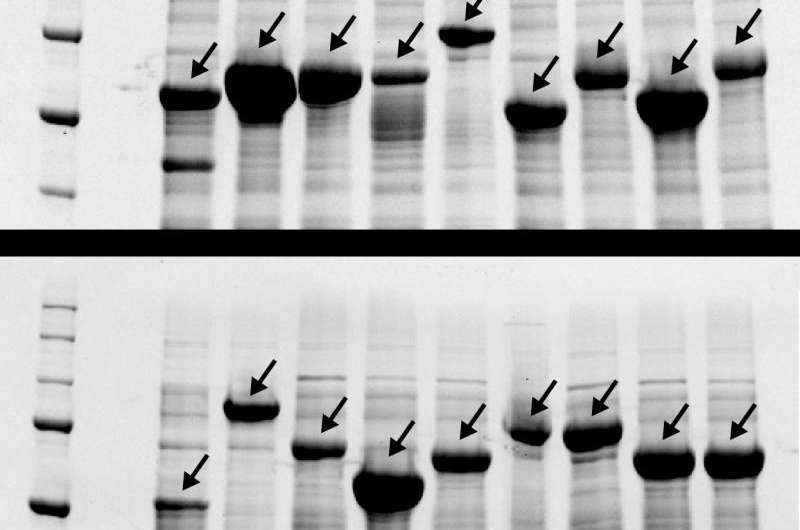
After writing the algorithm, Tang put it to the test. He created a laundry list of 19 popular repetitive polypeptides that are currently being studied in laboratories around the world. After passing the codes through the program, he sent them for synthesis by commercial biotechnology outfits—a task that would be impossible for any one of the original codes.
Without the help of commercial technology, researchers spend months building the DNA that cells use to produce the proteins being studied. It's a tedious, repetitive task—not the most attractive prospect to a young graduate student. But if the new program worked, the process could be reduced to a few weeks of waiting for machines to deliver the goods instead.
When Tang received his DNA, they each were introduced into living cells to produce the desired polypeptide as hoped.
"He made 19 different polymers from the field in one shot," said Chilkoti. "What probably took tens of researchers years to create, he was able to reproduce in a single paper in a matter of weeks."
Chilkoti and Tang are now working to make the new computer program available online for anybody to use through a simple web form, opening a new area of synthetic biology for all to explore.
"This advance really democratizes the field of synthetic biology and levels the playing field," said Tang. "Before, you had to have a lot of expertise and patience to work with repetitive sequences, but now anyone can just order them online. We think this could really break open the bottleneck that has held the field back and hopefully recruit more people into the field."
More information: Nicholas C. Tang et al. Combinatorial codon scrambling enables scalable gene synthesis and amplification of repetitive proteins, Nature Materials (2016). DOI: 10.1038/NMAT4521
Journal information: Nature Materials
Provided by Duke University