Exact pol(e) position—precisely where the polymerase is changed
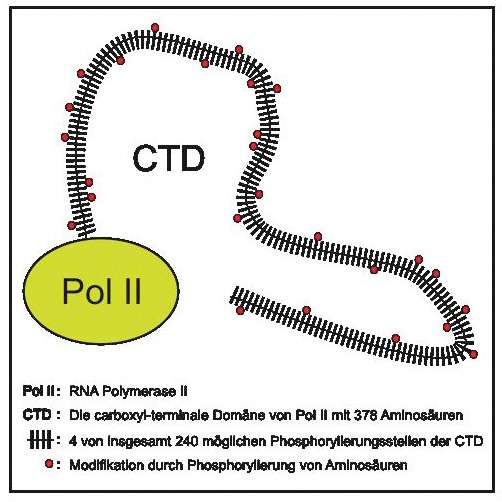
Scientists at the Helmholtz Zentrum München, working with colleagues from the Ludwig-Maximilians-Universität München, have developed a method for the thorough analysis of protein modifications. They mapped the phosphorylation sites of the RNA polymerase II enzyme, which is responsible for expressing our genes. The results have now been published in the Molecular Cell scientific journal.
The contents in our genetic information are actually silent (meaning inactive) and first have to be made to "speak". Like the read head in a tape recorder, RNA polymerase II, Pol II for short, runs over the DNA (tape) and transcribes the genetic and epigenetic information into RNA. In order to keep the enzyme from working randomly, however, it is dynamically modified at many different points in order to control its activity depending on the situation.
"Phosphorylation makes it possible to influence the activity of the enzyme at 240 different sites," explains Prof. Dirk Eick, the study's last author and head of the Research Unit Molecular Epigenetics at Helmholtz Zentrum München. Together with colleagues from the Biomedical Center and Gene Center of the Ludwig-Maximilians-Universität München and the Max Planck Institute for Biophysical Chemistry in Göttingen, Germany, he and his team have developed a method for simultaneously examining all 240 sites in Pol II.
"The trick is a combination of genetic and mass spectrometric methods," reveals first author Dr. Roland Schüller. "By producing genetically modified variants of the regions in question, we can examine each individual phosphorylation site with a mass spectrometer." This allows the researchers to determine exactly how and precisely where certain enzymes that influence phosphorylation act. The scientists also successfully compared the Pol II modification patterns in humans and in yeast.
"The regulation of the transcription of genes by Pol II is an elementary process in life and gene regulation deviations are the basis for many human disorders," study leader Eick explains the work's background. "Research into the phosphorylation pattern at certain times during the transcription cycle is therefore necessary in order to be able to gain an understanding of the underlying mechanisms of gene regulation at the transcription level sometime in the future."
More information: Schüller, R. et al. (2015). Heptad-specific Phosphorylation of RNA Polymerase II CTD, Molecular Cell, DOI: 10.1016/j.molcel.2015.12.003
Journal information: Molecular Cell
Provided by Helmholtz Association of German Research Centres