Down to the atom: Bacterial skeleton in close-up
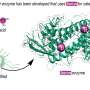
For a long time, bacteria were considered to be primitive structures, and it was not until the most modern of imaging techniques were used that their fine inner structure was discovered. The Berlin biophysicist Adam Lange has now succeeded in zooming in right up close: With the aid of a new technique of structural elucidation, he was able to show the basic building block of a bacterial skeleton down to atomic detail. The bactofilin investigated by his team only occurs in bacteria and may thus become a starting point for new antibiotics.
Bactofilin, discovered just five years ago, is found among other things in the bacterium Helicobacter pylori, which is responsible for the majority of gastric ulcers. Whereas it used to be thought that bacteria do not have a stabilising cytoskeleton, today we know that these microorganisms are in fact full of complex architectures, similar to the larger and in evolutionary terms more modern cells of plants and animals. Bactofilin gives Helicobacter pylori its typical screw-shaped form, thanks to which the bacterium can bore into the protective mucous layer of the inner wall of the stomach. The individual bactofilin molecules polymerise spontaneously in the interior of the bacteria to form the finest of fibres and higher order structures. An unusual structural motif plays a role here, as already discovered by the team of Adam Lange at the Leibniz-Institut für Molekulare Pharmakologie (FMP) in research work published at the beginning of this year. The proposed beta-helix fold had never before been found in a cytoskeleton. The bactofilin molecules are similar in form to spiral noodles with six twists, and in the process of polymerisation they accumulate together into long, extremely thin fibres.
The investigation of such fibre proteins is a challenge for structural biologists, as they can neither be dissolved in liquid nor can they be crystallised out, as is necessary for the commonly used methods. The two first authors of the paper, Chaowei Shi and Pascal Fricke, therefore used the relatively modern solid-state NMR, applying a new form of this technique further developed at the FMP that enables a particularly high resolution. NMR is the abbreviation for "nuclear magnetic resonance". It is based on the property of some atomic nuclei in a strong external magnetic field themselves to be turned into small magnets. On the basis of their characteristic resonance with radio waves, complicated calculation methods can be used to determine the position of the atoms within molecules. The special thing about solid-state NMR is that the sample is rotated very rapidly in the magnetic field in order to simulate the movements of dissolved molecules.
Since the exact form of the bactofilin building blocks and their chemical properties are now known, it is possible to search for small molecules that interfere with the polymerisation of the fibres. In this way, it might be possible to develop active substances that can specifically kill certain bacteria. The bactofilin fibres not only pass through the interior of Helicobacter – in the harmless Caulobacter crescentus the fibres even form tightly interwoven mats. These mats are the foundation for a long stalk with which the bacteria can attach to surfaces or take up nutrients.
"All processes in living organisms are ultimately driven by proteins, and we have to know their structures in order to understand how they function," says Adam Lange. The biophysicist is one of the world's leading experts in the field of solid-state NMR, but in future he wants to push ahead with the combination of different techniques. "Impressive breakthroughs have also been achieved in the field of cryo-electron microscopy over the past few years, and we want to establish co-operations here," says Lange. "If one wants to understand protein structures in all their dimensions and details, experts must not work on their own in isolation, but rather we must integrate the modern powerful techniques into joint projects."
More information: C. Shi et al. Atomic-resolution structure of cytoskeletal bactofilin by solid-state NMR, Science Advances (2015). DOI: 10.1126/sciadv.1501087
Journal information: Science Advances
Provided by Forschungsverbund Berlin e.V. (FVB)