Search engine for more accurate and fast recognition of metabolites
Potential applications for the machine-learning based method include anti-doping work, drug control by the Customs and crime scene investigation.
Researchers from Aalto University and the University of Jena in Germany have developed a search engine named CSI:FingerID that identifies metabolites from tandem mass spectrometry measurements with an accuracy more than 150 per cent higher than its rivals, which may make the work of researchers in life and medical sciences easier. The study was recently published in the highly regarded PNAS journal.
Metabolites are small molecules, such as sugars, fatty acids and amino acids that, among other things, serve as sources of energy in the cells and as building materials for cell walls. For researchers they are, as it were, traces of the functioning and status of cells.
'There are lots of metabolites, from hundreds of thousands to millions, and they all look a bit alike. In our study, we constructed a model that relies on machine learning. The molecular structures it predicts can be used in much the same way as search results from the Google search engine,' explains Professor Juho Rousu from Aalto University.
Molecule fingerprints
The tandem mass spectrometer used in the study is an instrument that splits molecules into fragments to measure their masses and relative abundances, or their mass spectrum. In the method developed by researchers from Aalto and Jena, a fragmentation tree is first computed from each spectrum included in the training data that describes for each fragment its parent, a larger fragment where it originated. Then, the researchers train the machine learning model using a large number of fragmentation trees and molecular properties or fingerprints that corresponds to each tree. When the spectrum of a new molecule is then given for the model, it predicts its probable fingerprints based on which a set of best-matching molecules is retrieved from the molecule database.
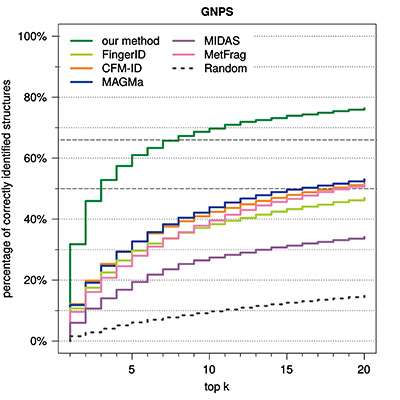
Depending on the type of the molecules, as much as 95 per cent of the searches currently return the correct search result among the top 10 matches. The accuracy of the identification improves as the volume of the data is increased. Currently, approximately 6,000 mass spectra have been used in building up the model. In an ideal situation, the machine-learning based search engine would always suggest the correct molecule as the first match, but this calls for a considerable increase in the data volume and further development of the methods.
The study could benefit researchers in life and medical sciences in particular. Potential future application areas include anti-doping work, drug control by the Customs and crime scene investigation.
Conducted in collaboration with a research group headed by Professor Sebastian Böcker of the University of Jena, the study serves as a good example of Aalto University's research that combines information technology with digital health.
More information: "Searching molecular structure databases with tandem mass spectra using CSI:FingerID." PNAS www.pnas.org/content/early/201 … /1509788112.abstract
Journal information: Proceedings of the National Academy of Sciences
Provided by Aalto University