Bacteria become 'genomic tape recorders'
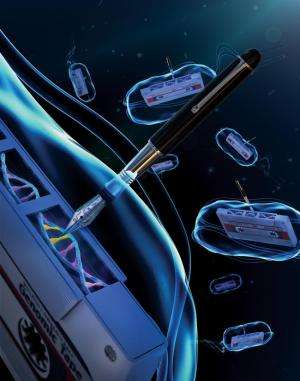
MIT engineers have transformed the genome of the bacterium E. coli into a long-term storage device for memory. They envision that this stable, erasable, and easy-to-retrieve memory will be well suited for applications such as sensors for environmental and medical monitoring.
"You can store very long-term information," says Timothy Lu, an associate professor of electrical engineering and computer science and biological engineering. "You could imagine having this system in a bacterium that lives in your gut, or environmental bacteria. You could put this out for days or months, and then come back later and see what happened at a quantitative level."
The new strategy, described in the Nov. 13 issue of the journal Science, overcomes several limitations of existing methods for storing memory in bacterial genomes, says Lu, the paper's senior author. Those methods require a large number of genetic regulatory elements, limiting the amount of information that can be stored.
The earlier efforts are also limited to digital memory, meaning that they can record only all-or-nothing memories, such as whether a particular event occurred. Lu and graduate student Fahim Farzadfard, the paper's lead author, set out to create a system for storing analog memory, which can reveal how much exposure there was, or how long it lasted. To achieve that, they designed a "genomic tape recorder" that lets researchers write new information into any bacterial DNA sequence.
Stable memory
To program E. coli bacteria to store memory, the MIT researchers engineered the cells to produce a recombinase enzyme, which can insert DNA, or a specific sequence of single-stranded DNA, into a targeted site. However, this DNA is produced only when activated by the presence of a predetermined molecule or another type of input, such as light.

After the DNA is produced, the recombinase inserts the DNA into the cell's genome at a preprogrammed site. "We can target it anywhere in the genome, which is why we're viewing it as a tape recorder, because you can direct where that signal is written," Lu says.
Once an exposure is recorded through this process, the memory is stored for the lifetime of the bacterial population and is passed on from generation to generation.
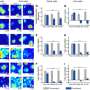
There are a couple of different ways to retrieve this stored information. If the DNA is inserted into a nonfunctional part of the genome, sequencing the genome will reveal whether the memory is stored in a particular cell. Or, researchers can target the sequences to alter a gene. For example, in this study, the new DNA sequence turned on an antibiotic resistance gene, allowing the researchers to determine how many cells had gotten the memory sequence by adding antibiotics to the cells and observing how many survived.
By measuring the proportion of cells in the population that have the new DNA sequence, researchers can determine how much exposure there was and how long it lasted. In this paper, the researchers used the system to detect light, a lactose metabolite called IPTG, and an antibiotic derivative called aTc, but it could be tailored to many other molecules or even signals produced by the cell, Lu says.
The information can also be erased by stimulating the cells to incorporate a different piece of DNA in the same spot. This process is currently not very efficient, but the researchers are working to improve it.
Bacterial sensors
Environmental applications for this type of sensor include monitoring the ocean for carbon dioxide levels, acidity, or pollutants. In addition, the bacteria could potentially be designed to live in the human digestive tract to monitor someone's dietary intake, such as how much sugar or fat is being consumed, or to detect inflammation from irritable bowel disease.
These engineered bacteria could also be used as biological computers, Lu says, adding that they would be particularly useful in types of computation that require a lot of parallel processing, such as picking patterns out of an image.
"Because there are billions and billions of bacteria in a given test tube, and now we can start leveraging more of that population for memory storage and for computing, it might be interesting to do highly parallelized computing. It might be slow, but it could also be energy-efficient," he says.
Another possible application is engineering brain cells of living animals or human cells grown in a petri dish to allow researchers to track whether a certain disease marker is expressed or whether a neuron is active at a certain time. "If you could turn the DNA inside a cell into a little memory device on its own and then link that to something you care about, you can write that information and then later extract it," Lu says.
More information: "Genomically encoded analog memory with precise in vivo DNA writing in living cell populations," by F. Farzadfard et al. Science, 2014.
Journal information: Science
Provided by Massachusetts Institute of Technology