Antibiotic resistance genes are essentially everywhere
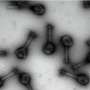
The largest metagenomic search for antibiotic resistance genes in the DNA sequences of microbial communities from around the globe has found that bacteria carrying those vexing genes turn up everywhere in nature that scientists look for them. The findings reported in Current Biology on May 8 add to evidence showing just how common and abundant those resistance genes really are in natural environments.
This big-picture, ecological view on a growing healthcare concern emphasizes the important relationship between antibiotic resistance in the clinic and environmental microbiology, the researchers say.
"While the environment is known to harbor antibiotic-resistant strains of bacteria, as proven by many preceding studies, we did not really know the extent of their abundance," says Joseph Nesme of the Université de Lyon in France. "The fact that we were able to detect antibiotic resistance genes at relatively important abundance in every environment tested is certainly our most striking result."
The researchers, including Nesme and senior author of the study Pascal Simonet, took advantage of the ever-growing reams of existing next-generation sequencing data that are freely available in public repositories together with information about antibiotic resistance genes found in pathogens infecting patients in the clinic.
"Our strategy was simply to use all these pre-existing data and combine them to answer more precisely the question of antibiotic resistance prevalence in the environment," Nesme says.
The scientists' analyses detected antibiotic resistance gene determinants in all 71 environments represented in the public data, including soil, oceans, and human feces. Samples collected from soil contained the most diverse pool of resistance genes, the authors found. The most common types of resistance uncovered were efflux pumps and other genes conferring resistance to vancomycin, tetracycline, or beta-lactam antibiotics, which are in common use in veterinary and human healthcare.
All this, and Simonet says they know that today's technologies are still unable to capture all of the diversity present in the environment. In other words, we're still missing part of the picture.
There is a very good reason microbes would be armed with antibiotic resistance genes, the researchers explain. After all, most antibiotics used in medicine are isolated from soil microorganisms, such as bacteria or fungi, in the first place. That means that the resistance genes were available long before humans put antibiotic drugs into use. Bacteria lacking them to start with can simply borrow them (via horizontal transfer of genes) from those that are better equipped.
Nesme and Simonet say the new findings should come as a plea for a broader ecological perspective on the antibiotic resistance problem.
"It is only with more knowledge on antibiotic resistance dissemination—from the environment to pathogens in the clinic and leading to antibiotic treatment failure rates—that we will be able to produce more sustainable antibiotic drugs," Nesme says.
More information: Current Biology, Nesme et al.: "Large-scale metagenomic-based study of antibiotic resistance in the environment." http://www.cell.com/current-biology/abstract/S0960-9822(14)00328-5
Journal information: Current Biology
Provided by Cell Press