Huntington proteins and their nasty 'social network'
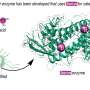
Researchers at the Buck Institute have identified and categorized thousands of protein interactions involving huntingtin, the protein responsible for Huntington's disease (HD). To use an analogy of a human social network, the identified proteins are like "friends" and "friends of friends" of the HD protein. The network provides an invaluable resource for identifying targets to treat the disease and has been used to implicate a particular signaling pathway involved in cell motility. HD is an incurable, fatal, inherited neurological disorder that causes severe degeneration of the nervous system.
The research appears in the March 7, 2014 edition of the Journal of Biological Chemistry and was chosen as the Paper of the Week. The Journal's editorial board members consider this study to be in the top 2% of those to be published this year in terms of significance and overall importance.
HD is caused by a mutation in the human HTT gene that results in an abnormal expansion and misfolding of the corresponding huntingtin protein. Buck researchers established an unprecedented large-scale interaction network for the huntingtin protein identifying 2,141 highly interconnected proteins that have over 3,200 interactions among them.
The work involved a close collaboration between Buck faculty members Robert E. Hughes, PhD, an expert in neurodegeneration, and Sean D. Mooney, PhD, who leads the Institute's bioinformatics program. Researchers analyzed protein interaction data generated at Prolexys Pharmaceuticals that identified more than 100 huntingtin interacting proteins (HIPs) and more than 2,000 proteins that interact with HIPs. "The damage caused by the mutant huntingtin protein radiates out through the cell, like a pebble dropped in a pond. In this case, the pond is filled with proteins that make up much of the cell," said Hughes. "We now have a handle on the detailed structure of a complex web of interactions that causes global dysfunction in cells resulting in degeneration of the brain."
Hughes said Mooney employed sophisticated computational methods which allowed researchers to comprehensively analyze the functions or so-called "jobs" of the proteins and networks and how they might be impacted by the huntingtin mutation. The investigators identified several pathways that were particularly conspicuous in the network. In particular, HD mutations impacting the RhoGTPase signaling pathway interfered with filopodia, the slender projections that cells use to direct movement and communicate with other cells. The data indicate that the HD mutation directly affects membrane dynamics, cell attachment and cell motility. Defects in these pathways can provide critical clues for how to best intervene in the disease with drugs.
Highlighting the collaboration, Hughes said, "This study demonstrates how the synergy between experimental and computational approaches can help unravel the nature of a complex disease such as HD." Mooney added, "Understanding and characterizing potentially functional HD protein interactions gives scientists new tools to connect genomic, genetic, proteomic and other molecular changes to identify the causes of this deadly disease. Bioinformaticians can add this dataset to their systems biology toolbox in the quest for interventions that can suppress the progression of HD."
More information: Journal of Biological Chemistry; Vol. 289, issue 10; PubMed PMID: 24407293.
Journal information: Journal of Biological Chemistry
Provided by Buck Institute for Age Research