How DNA damage affects Golgi—the cell's shipping department
In studying the impact of DNA damage on the Golgi, a research team from the University of California, San Diego School of Medicine and the Ludwig Institute for Cancer Research have discovered a novel pathway activated by DNA damage, with important consequences for the body's cellular response to chemotherapy.
Standard cancer treatments, including many chemotherapy drugs and radiation therapy, act on cells by causing DNA damage. In many cancer cells, DNA damage turns on signaling pathways that lead to cell death – the basis of the use of these treatments for cancer.
A better understanding of the signaling pathways that are activated in cells in response to DNA damage, and the influence they exert to determine the fate of the cell to live or die, ultimately could lead to more effective use of these DNA damaging agents to treat cancer.
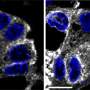
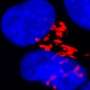
A study published in the January 30, 2014 issue of the journal Cell – led by Seth Field, MD, PhD, associate professor of medicine at UC San Diego School of Medicine – demonstrates that DNA damage triggers dramatic reorganization of the Golgi. The Golgi serves as the cell's processing center for the exportation of proteins, lipids and other large molecules to their final destinations outside of the cell. The researchers showed that, in mammalian cells, DNA damage triggers the Golgi to fragment and disperse throughout the cell.
In 2009, the research team had discovered a three-way interaction between a particular Golgi protein, GOLPH3, a lipid signaling molecule, PtdIns(4)P and a contractile protein, MYO18A. The link between the three applies a tensile force required for effective formation of the tubules and vesicles necessary for extracellular transportation.
Later screening identified GOLPH3 as an oncogene overexpressed in many human cancers, which can transform cells into tumorous cells. This study shows that common cancer therapeutic agents, by triggering DNA damage, activate GOLPH3.
Examining the mechanism of Golgi dispersal, the researchers discovered that Golgi dispersal in response to DNA damage involves a novel signaling pathway that directly links the DNA damage response to the Golgi.
The study also showed that the DNA damage-activated protein kinase, DNA-PK, directly modifies the Golgi protein GOLPH3 by phosphorylation on a specific site. This, in turn, enhances the interaction of GOLPH3 with MYO18A, increasing the tensile force applied to the Golgi, causing Golgi dispersal.
Interfering with Golgi dispersal after DNA damage by depletion of any of the components of this pathway – including DNA-PK, GOLPH3, or MYO18A – resulted in enhanced cell killing by DNA damaging agents. The scientists concluded that this pathway is normally required to allow cells to survive DNA damage.
"We further found that overexpression of GOLPH3, as is seen in human cancers, protects cells from killing by DNA damaging agents," said Field.
Identification of such a Golgi response reveals an unexpected pathway through DNA-PK, GOLPH3 and MYO18A that regulates cell survival following DNA damage, Field added. "This unappreciated feature of the cellular DNA damage response plays a significant role in determining cell survival."
Journal information: Cell
Provided by University of California - San Diego