Probe opens new path for drug development against leading STD
Biochemical sleuthing by an Indiana University graduate student has ended a nearly 50-year-old search to find a megamolecule in bacterial cell walls commonly used as a target for antibiotics, but whose presence had never been identified in the bacterium responsible for the most commonly reported sexually transmitted disease in the United States.
For decades researchers had searched for peptidoglycan—a mesh-like polymer that forms the cell wall in diverse bacteria—in the bacterial pathogen Chlamydiae in hopes of studying the megamolecule's structure and synthesis as a path to drug development against a class of bacteria responsible for 1 in 10 cases of pneumonia in children and over 21 million cases of the blindness-causing disease trachoma.
So when IU graduate student Erkin Kuru and a team of researchers that included IU biologist Yves Brun and chemist Michael S. VanNieuwenhze announced discovery in October 2012 of the first direct and universal approach for labeling peptidoglycan, one of the first places Kuru sought to put his new set of designer chemicals to work was against Chlamydiae.
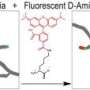
"People had been trying for about 50 years to identify peptidoglycan in Chlamydiae but had failed even though there was a lot of indirect evidence pointing to its existence," said Kuru, a native of Istanbul, Turkey, doing graduate studies in the IU Bloomington College of Arts and Sciences' Interdisciplinary Biochemistry Program. "We immediately thought to put our new chemical tagging method using fluorescent D-amino acids (FDAAs) to work in an effort to uncover the molecular signature of peptidoglycans in Chlamydiae."
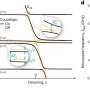
The new method uses nontoxic D-amino acid-based fluorescent dyes to label sites where peptidoglycan is synthesized, allowing for fine spatiotemporal tracking of cell wall dynamics. Antibiotics including the penicillins, the cephalosporins and vancomycin all target the peptidoglycan assembly site, yet resistance of bacteria to these antibiotics is on the rise.
Kuru quickly sought the collaborative support of one of the world's leading Chlamydiae researchers, Anthony Maurelli of the U.S. government's Uniformed Services University of the Health Sciences, and forwarded the new FDAA probes to Maurelli's laboratory in Bethesda, Md.
"When the FDAAs failed to label peptidoglycan in Chlamydiae, my postdoc George Liechti and I were obviously disappointed. But we really believed that Chlamydiae made peptidoglycan so we encouraged Erkin to come up with a new labeling method," Maurelli said. "We hypothesized that enzymes unique to synthesizing peptidoglycan in Chlamydiae may not have recognized the FDAA probes, so we needed Erkin to develop an alternative labeling strategy."
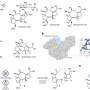
"It turned out the strategy Erkin needed was already in his tool box," VanNieuwenhze said. "As an alternative labeling strategy, we had synthesized a different type of probe, this time mimicking the last two amino acids of the peptidoglycan peptide sequence."
The IU team, with chemistry support from graduate student Edward Hall and postdoctoral researcher Alvin Kalinda, linked the two amino acids (D-alanine-D-alanine, or dipeptide) to chemical groups that could be selectively functionalized with fluorescent probes, after their incorporation into peptidoglycan, by a very specific chemical reaction called click chemistry. The clickable dipeptides were used and validated with a number of other bacterial species to show they were incorporated in peptidoglycan, and the new probes were forwarded to Maurelli's team.
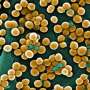
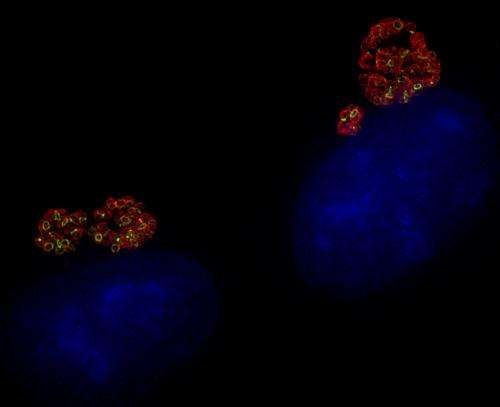
When the new dipeptide probes were applied to cells infected with Chlamydia trachomatis, it worked immediately, said Liechti, a postdoctoral researcher in Maurelli's lab who conducted the experiments.
"The chlamydia inside the infected cells lit up with green fluorescent peptidoglycan rings in the middle of the red-stained bacterial cells," Liechti said. "It was amazing."
The ability to detect the peptidoglycan will make it possible to solve the mystery of why Chlamydiae peptidoglycan has remained undetected for so long.
"Obviously, it must be structurally different from the peptidoglycan of most bacteria, and understanding the basis for this difference will help develop strategies to fight this dangerous pathogen," Brun said.
The new research, "A new metabolic cell wall labeling method reveals peptidoglycan in Chlamydia trachomatis," by Kuru, Brun, VanNieuwenhze, graduate student Edward Hall and former postdoctoral researcher Alvin Kalinda, all affiliated with IU, and Liechti and Maurelli, was published today, Dec. 11, in Nature.
More information: A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis, DOI: 10.1038/nature12892
Journal information: Nature
Provided by Indiana University