How scavenging fungi became a plant's best friend
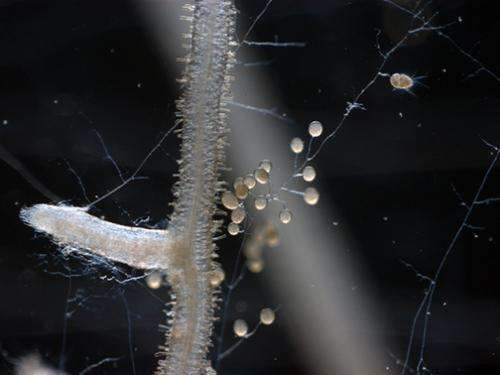
Glomeromycota is an ancient lineage of fungi that has a symbiotic relationship with roots that goes back nearly 420 million years to the earliest plants. More than two thirds of the world's plants depend on this soil-dwelling symbiotic fungus to survive, including critical agricultural crops such as wheat, cassava, and rice. The analysis of the Rhizophagus irregularis genome has revealed that this asexual fungus doesn't shuffle its genes the way researchers expected. Moreover, rather than having lost much of its metabolic genes, as observed in many mutualistic organisms, it has expanded its range of cell-to-cell communication genes and phosphorus-capturing genes.
A team led by the French National Institute for Agricultural Research (INRA) and including researchers from the Department of Energy Joint Genome Institute (DOE JGI) reported the complete genome of R. irregularis (formerly Glomus intraradices) in a paper published online November 25 in the journal Proceedings of the National Academy of Sciences (PNAS). The fungus is a member of the Glomeromycota family and frequently colonizes many plants important to agriculture and forestry. Glomeromycota, also called arbuscular mycorrhizal fungi (AMF), play a vital role in how phosphorus and carbon cycles through the atmosphere and land-based ecosystems, but exactly how it does this vital job is poorly understood.
"This is the first sequenced genome of arbuscular mycorrhizae, the type that is dominant on the planet," said Igor Grigoriev, one of the senior authors on the paper and lead for the Fungal Genomics Program at the DOE JGI.
It was a long hard road to a sequenced arbuscular mycorrhizal fungus. In 2006, shortly after the DOE JGI sequenced the first tree genome, Populus trichocarpa, it became apparent that it took a village (of other organisms) to raise a poplar tree. Researchers Jerry Tuskan of Oak Ridge National Laboratory and Francis Martin of INRA, recommended that the assembly of Populus-associated fungi and bacteria be sequenced to inform research on perennial plant growth, ecosystem function and plant microbe interactions. This long passage is outlined in an earlier publication in New Phytologist. Rhizophagus irregularis, is the next in this linage to be released by the DOE JGI, it follows the ectomycorrhizal fungal symbiont Laccaria, the poplar rust pathogen Melampsora, and dozens of bacterial genomes.
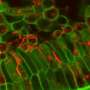
A relic of fungal evolution, AMF diverged early on from other forms of fungus. They form dense clusters of branched structures—called arbuscules—in root cells, much like a tight, many-fingered handhold. The arbuscules are the main route of nutrient exchange between plants and fungi. Unable to live on their own, AMF are entirely dependent on their plant hosts for the sugars they need for food. They have carefully established their relationship with host plants, keeping them alive while sapping nutrients from them.
But AMF are also adept at capturing phosphorus from the soil and making it available for their hosts. Phosphorus, a critical element for cellular function, is otherwise difficult to extract from the soil and is often the limiting factor for how quickly a plant grows.
Scientists theorize that the benefits these fungi provided enabled ancient plants to evolve during the Paleozoic era, about 250 to 500 million years ago. Over time, plants adapted their essentially rootless primordial form and developed deeper and stronger roots to take advantage of the nutrients that underground AMF fed them. In exchange, plants provided nutrients the fungi couldn't obtain themselves.
Analysis of the R. irregularis genome also revealed several surprising details. The research team found that the genome is among the largest fungal genomes sequenced, weighing in at 153 million base pairs (Mb). For comparison, the button mushroom (Agaricus bisporus), also sequenced and published by the DOE JGI, has a genome of about 30 Mb. Through several generations, portions of R. irregularis's genome were duplicated, invaded by repeated transposable elements, famously known as 'jumping genes'. Unlike many other fungi, R. irregularis seems to lack mechanisms that can keep these transposable elements from running amok.
"Among the expanded portions of its genome, R. irregularis had several genes for phosphorus metabolism, which are probably responsible for its large appetite for phosphorus," said Francis Martin, one of the senior authors on the paper and lead for the Cluster of Excellence, Advanced Research on the Biology of Tree and Forest Ecosystems (ARBRE) at the INRA. "They also have an abundance of genes for communication between cells via signaling proteins, including small secreted effectors highly expressed during symbiosis. Plant roots send out a plethora of chemical signals and these genes probably help AMF interact with plants, picking up the signals plants pump out."
Another surprise for the research team was in the genes that govern metabolism. "Obligate parasites often have broken metabolism, missing some genes in critical metabolic pathway which make them dependent on their host," Grigoriev said. "We did not find such genes here." R. irregularis has retained much of its metabolic machinery, unlike many other obligate parasitic organisms. It leads a double-life, extracting minerals from the soil while still living in harmony with its host plant.
Though it has nearly 30,000 protein-encoding genes, R. irregularis has also lost hundreds of genes as a result of its close association with plants. For example, it can't make most of the toxins other plant-interacting fungi release, probably, the researchers speculate, to avoid setting off the host plant's immune system. It has also cast off most of its genes for breaking down plant cell walls, a critical ability for free-living fungi that feed off dead organic matter in soils.
Teasing apart the complex relationship between soil fungi and plants is likely to have an impact on improving biofuel production from plant biomass. "Through analysis of this and other mycorrhizal genomes, we can help to better understand interactions and conditions critical for a sustainable growth of bioenergy plants, but also staple crops, a prerequisite to help feeding the world," said Martin.
More information: Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis, www.pnas.org/cgi/doi/10.1073/pnas.1313452110
Journal information: Proceedings of the National Academy of Sciences , New Phytologist
Provided by DOE/Joint Genome Institute