Molecular structure reveals how the antibiotic streptomycin works
(Phys.org) —Streptomycin was the first antibiotic developed to treat tuberculosis yet until recently, scientists did not completely understand how it works at the molecular level. They knew that streptomycin blocks a critical process, the synthesis of proteins by ribosomes leading to bacterial cell death, but certain details of the interaction remained undiscovered. At Brookhaven National Laboratory's National Synchrotron Light Source, researchers have used x-ray crystallography to complete the picture.
Streptomycin is a member of a family of antibiotics that work by interrupting the function of bacteria cells' ribosomes, the complex molecular machines that create proteins by linking amino acids together. Ribosomes, a major target for antibiotics that work by inhibiting the synthesis of proteins, have two main parts or "subunits."
The larger subunit does the protein building, guided by a type of RNA called messenger RNA (mRNA), which binds to it. The small subunit "reads" the mRNA and selects the matching transfer RNA (tRNA) molecule, which selects and delivers the next amino acid to the ribosome. This is where streptomycin plays a role. It binds close to the small subunit, causing it to severely misread the sequence. This results in the synthesis of random proteins, which ultimately kills the bacteria. But how this misreading occurred remained a mystery, until a recent study by researchers from Brown University and the multi-institution Northeastern Collaborative Access Team at Argonne National Laboratory (managed by Cornell University).
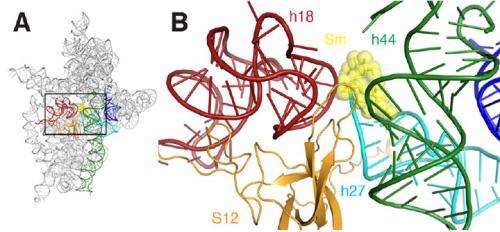
By creating a crystal – an ordered structure of identical units – of the small ribosomal subunit bound to mRNA in the presence of streptomycin, the researchers generated several detailed "snapshots" that revealed key molecular-level details of the interaction, ultimately showing how streptomycin impairs the function of the subunit. At NSLS beamline X25, they used a technique called x-ray crystallography, in which a beam of x-rays is aimed at the crystal, interacts with the molecules, and yields an intricate diffraction pattern. From the pattern, with the help of computer software, the group constructed visual representations of the subunit-mRNA-streptomycin complex.
In short, the researchers could "see" for the first time the subtle ways in which streptomycin distorts the structure of the subunit's decoding site, causing it to incorrectly read the mRNA. For example, streptomycin binding reduces the distance between two of the many helices that make up the subunit's molecular structure. This is particularly significant because these helices form the actual decoding site, and decoding only takes place properly if these elements are oriented exactly right with respect to the mRNA and the selected tRNA. Streptomycin binding also induces a change in the relationship between one of these two helices and a third helix, causing one to retract away from the other or "disengage."
The end result of all of these slight alterations is that streptomycin destabilizes binding between the subunit and the "correct" tRNA while simultaneously stabilizing the binding of the subunit to the "wrong" tRNA, thereby effectively removing the discrimination between the correct and the wrong tRNA. This causes havoc in the bacterial supply chain for new proteins, disrupting the bacteria reproduction and life cycle.
"Our structural studies revealed that the streptomycin induces surprisingly large distortions in the bacterial ribosome, which help us understand how this antibiotic interferes with protein synthesis in bacteria," said lead researcher Gerwald Jogl, an associate professor of biology in Brown's Molecular Biology, Cell Biology & Biochemistry Department. "Continuing from our current findings, we are now studying how mutations in bacterial ribosomes can counteract these structural rearrangements and enable bacteria to survive the otherwise lethal action of streptomycin."
This research was published in the January 15, 2013 edition of Nature Communications, under the title "A structural basis for streptomycin-induced misreading of the genetic code." Support came from the National Institutes of Health and the Department of Energy.
More information: www.nature.com/ncomms/journal/ … full/ncomms2346.html
Journal information: Nature Communications
Provided by Brookhaven National Laboratory