Jekyll and Hyde: motion may explain similar enzymes' divergence
One enzyme regulates the body's insulin receptor, ensuring energy needed for function and survival. The other enables a bacterium to wreak havoc in the form of bubonic plague.
"Their purposes in life couldn't be more different," says Utah State University chemist Alvan Hengge.
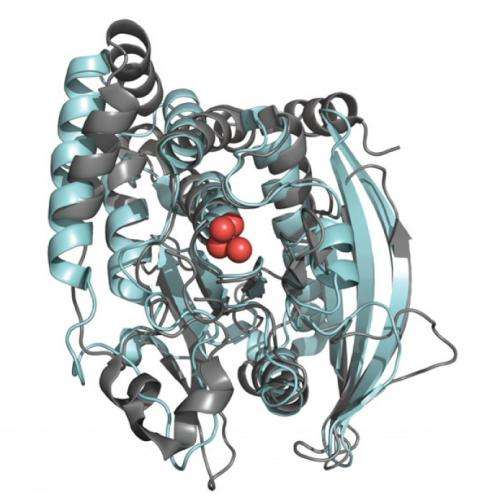
From a molecular standpoint, the two enzymes appear virtually identical. Yet, they catalyze the same reaction at very different rates.
With colleagues Sean Whittier and Patrick Loria of Yale University, Hengge, who has puzzled over this 'Jekyll-Hyde' conundrum for several years, published findings that may explain the dissimilarities in the Aug. 23, 2013, issue of Science. The research was supported by funding from the National Institutes of Health and the National Science Foundation.
"We've known since the 1990s that molecular motions in the class of enzymes called protein tyrosine phosphatases are crucial for their optimal function," says Hengge, professor and head of USU's Department of Chemistry and Biochemistry. "What we're discovering is the speed of these motions is different among PTPs and is the reason some of them are much faster catalysts than others."
Using nuclear magnetic resonance, the team compared the active-site loop motions in two structurally similar enzymes. One, PTP1B, is the human phosphatase involved in regulation of the insulin receptor, as well as leptin and epidermal growth factor signaling. The other, YopH, is the virulence factor from the bacterium Yersinia, an invasive pathogen that causes life-threatening infection.
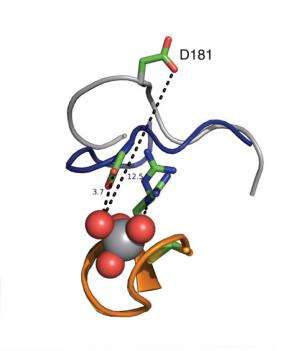
"In the molecular structure of each of these enzymes, there is a loop of the protein that must close after the substrate binds to catalyze the chemical reaction," Hengge says. "PTP1B's loop moves relatively slowly. In contrast, YopH's loop seems hyperactive; moving about 30 times faster."
From an evolutionary standpoint, he says, the difference makes sense.
"Slower PTP1B motions likely reflect the enzyme's physiological role as a tight regulator of cellular processes," Hengge says. "Its turnover rate has to meet the biological requirements of the cell. Speed is not everything."
In contrast, the speedy YopH enzyme plays no regulatory role.
"The faster loop motion and enzymatic activity enables YopH to facilitate Yersinia infection that disrupts metabolic pathways and allows the bacteria to move on to the next host," Hengge says.
More information: "Conformational Motions Regulate Phosphoryl Transfer in Related Protein Tyrosine Phosphatases," by S.K. Whittier et al. Science, 2013.
Journal information: Science
Provided by Utah State University