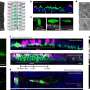
Minus environment, patterns still emerge: Computational study tracks E. coli cells' regulatory mechanisms
Environment is not the only factor in shaping regulatory patterns—and it might not even be the primary factor, according to a new Rice University study that looks at how cells' protein networks relate to a bacteria's genome.
The Rice lab of computer scientist Luay Nakhleh reported in the Proceedings of the National Academy of Sciences that when environmental factors are eliminated from an evolutionary model, mutations and genetic drift can give rise to the patterns that appear. They studied changes that show up in regulatory networks that determine the organism's characteristics.
Nakhleh and lead author Troy Ruths, a Rice graduate student, said their work is an important step toward understanding Cis-regulatory networks (CRNs), which play a dominant role in cells' information processing systems. Cis—a Latin word for "on the same side as"—regulators are regions of DNA (or RNA) that regulate the expression of genes located on the same DNA molecule.
The researchers generated 1,000 computer models of random Escherichia coli regulatory networks and watched them evolve through millions of generations. However, they did not program into the models environmental factors that might have prompted change through natural selection. Their results supported other studies that suggested networks might evolve spontaneously through mutation, recombination, gene duplication and genetic drift.
Their "neutral evolutionary" approach sidestepped one taken by other researchers who, Nakhleh said, have tried to understand cellular protein networks by looking at motifs. These motifs are short sequences called subgraphs in the network that appear more frequently than is expected by chance alone. "Biological networks are complex systems, and the community has responded by developing lots of mathematical and sophisticated computational analysis tools to understand these networks," he said. Those researchers argued the emergence and conservation of these regulatory motifs were largely due to adaptation to environment; the Rice researchers argued that isn't necessarily so.
Nakhleh said he and Ruths decided to tie what scientists now know about the genome – the entire collection of an organism's DNA – to the evolution of such networks.
"Instead of jumping directly to the network, where we don't understand much, we decided to look back at our broad knowledge about the genome and link it to these networks," he said. "In this paper, we zoomed in on the issue of how much of what we see in the network is a result of neutral evolution, where there's no selection involved. How much of what we are seeing is a side effect, so to speak, of random mutations and genetic drift?"
The wealth of genomic data available for E. coli encouraged the Rice researchers to build a sophisticated model that matched Cis-regulatory networks to their related DNA. "If there is any model in the prokaryotic world that has been studied well and has data, it's E. coli," Nakhleh said.
Their conclusion, put simply by the paper, is that "neutral evolution acting on genomic properties" can indeed explain bacterial regulatory patterns.
"There are two sides to the paper," Nakhleh said. "One is that many of these motifs have nothing adaptive in their origin. They emerge because mutation is a random process.
"The second and, I think, more powerful part of the story is that for the first time, the extent of neutrality in a network has been quantified. … Our model will never be able to tell you, 'I can rule out adaptation from this.' What we are saying is that you do not need to invoke adaptation to explain what you are seeing.
"Now we can start to understand how changes at the genome level can result in how these networks form, what some researchers are calling the 'design principles' behind these networks," Nakhleh said. "I don't think there is anything being designed here, so to speak. Patterns emerge in response to mutations; genetic drift and selection then affect the frequencies of these patterns. We showed that genetic drift can explain much of these frequencies."
More information: www.pnas.org/content/110/19/7754.full
Journal information: Proceedings of the National Academy of Sciences
Provided by Rice University