Cybernetic model developed to predict Shewanella metabolic behavior
To further the quest to harness microbes for beneficial uses, scientists from Pacific Northwest National Laboratory and Purdue University developed a promising computational tool for analyzing microbial flux distribution and metabolic engineering. They used the Lumped Hybrid Cybernetic Model (L-HCM), developed by Purdue researchers Dr. Hyun-Seob Song and Dr. Doraiswami Ramkrishna, to predict and simulate the metabolic dynamics of Shewanella oneidensis MR-1 during aerobic growth in a bioreactor.
In this use, L-HCM predicted experimentally determined dynamic metabolic shifts, a capability well beyond the scope of constraint-based modeling approaches. The researchers established procedures for modeling microbial metabolism during growth on different carbon compounds. Their research also provides the basis for modeling S. oneidensis under more complex environments.
This approach is unique in that it is able to model metabolic dynamics. S. oneidensis is of particular interest to scientists because of its capacity for extracellular electron transfer, which enables it to use a range of electron acceptors, including manganese and iron oxides, to generate energy. This makes it an attractive tool for bioremediation, microbial fuel cell applications, and biofuel production.
The microbe exhibits an intriguing growth pattern in the aerobic bioreactor by sequentially consuming substrate (lactate) and then metabolic by-products (pyruvate and acetate). What makes modeling this process so difficult is the complex growth pattern of Shewanella in batch culture, which scientists think is driven by its complex regulatory circuitry.
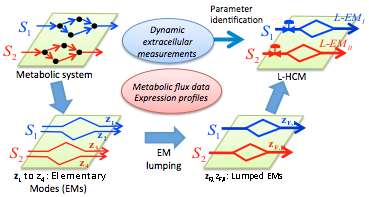
The ability to exploit the biotechnological potential of S. oneidensis MR-1 will benefit greatly from metabolic modeling and simulations that not only promote fundamental understanding, but also point to new strategies for performance improvements. To do so, a model must provide reliable descriptions of key system properties-in particular, the network of metabolic pathways and their dynamic regulation. Until now, no models could incorporate both of those components.
"The L-HCM provides an appropriate framework for a detailed network-based dynamic model," said Dr. Alex Beliaev, PNNL scientist and principal investigator on PNNL's Biofuels Science Scientific Focus Area (BSFA). "It considers both structural and functional components of metabolic systems and is capable of predicting complex organismal response without over-parameterizing the model."
Song and Ramkrishna originally demonstrated the L-HCM's ability to predict complex dynamic response of E. coli to environmental changes and genetic perturbation. In the current study, the model is used to investigate the aerobic growth of S. oneidensis MR-1 on lactate.
The L-HCM accurately reproduces exacting dynamic metabolic shifts and provides reasonable estimates of energy requirement for growth. Flux distributions in S. oneidensis MR-1 predicted by the L-HCM compare very favorably with 13C-metabolic flux analysis results reported in the literature. The model's predictive accuracy is enhanced by incorporating measurements of only a few intracellular fluxes, in addition to extracellular metabolites.
The research team first developed a general protocol for constructing an L-HCM from data showing sequential consumption of different alternative carbon sources. The resulting model demonstrated the ability to correctly predict growth dynamics and changes in extracellular flux distributions with time. The L-HCM results were compared with traditional constraint-based model approaches, as well as with their own experimental data and those available in the literature.
The researches found that modeling cellular regulation with cybernetic control laws provides an efficient route to describing the complex dynamic behavior of S. oneidensis MR-1 particularly when considering large-scale metabolic networks.
More information: Song, H. et al. 2013. Dynamic Modeling of Aerobic Growth of Shewanella oneidensis. Predicting Triauxic Growth, Flux Distributions and Energy Requirement for Growth. Metabolic Engineering 15:25-3. DOI:10.1016/j.ymben.2012.08.004
Provided by Pacific Northwest National Laboratory




















