New method for uncovering side effects before a drug hits the market
Side effects are a major reason that drugs are taken off the market and a major reason why patients stop taking their medications, but scientists are now reporting the development of a new way to predict those adverse reactions ahead of time. The report on the method, which could save patients from severe side effects and save drug companies time and money, appears in ACS' Journal of Chemical Information and Modeling.
Yoshihiro Yamanishi and colleagues explain that drug side effects are a major health problem—the fourth-leading cause of death in the U.S.—which by some estimates claim 100,000 lives every year. Serious side effects are the main reason why existing drugs must be removed from the market and why pharmaceutical companies halt development of new drugs after investing millions of dollars. Current methods of testing for side effects are costly and inaccurate. That's why the scientists sought to develop a new computer-based approach to predicting possible side effects.
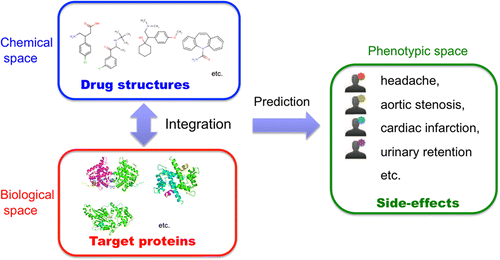
They show the usefulness of their proposed method on simultaneous prediction of 969 side effects of 658 drugs that already are in wide medical use. The method is based on knowledge about chemical and biological information about ingredients in these medications. They also used the approach to identify possible side effects for many uncharacterized molecules. Based on that work, the scientists conclude that the new method could be helpful in uncovering serious side effects early in the development and testing of new drugs, avoiding costly investment in medications unsuitable for marketing.
More information: Drug Side-Effect Prediction Based on the Integration of Chemical and Biological Spaces, J. Chem. Inf. Model., 2012, 52 (12), pp 3284–3292. DOI: 10.1021/ci2005548
Abstract
Drug side-effects, or adverse drug reactions, have become a major public health concern and remain one of the main causes of drug failure and of drug withdrawal once they have reached the market. Therefore, the identification of potential severe side-effects is a challenging issue. In this paper, we develop a new method to predict potential side-effect profiles of drug candidate molecules based on their chemical structures and target protein information on a large scale. We propose several extensions of kernel regression model for multiple responses to deal with heterogeneous data sources. The originality lies in the integration of the chemical space of drug chemical structures and the biological space of drug target proteins in a unified framework. As a result, we demonstrate the usefulness of the proposed method on the simultaneous prediction of 969 side-effects for approved drugs from their chemical substructure and target protein profiles and show that the prediction accuracy consistently improves owing to the proposed regression model and integration of chemical and biological information. We also conduct a comprehensive side-effect prediction for uncharacterized drug molecules stored in DrugBank and confirm interesting predictions using independent information sources. The proposed method is expected to be useful at many stages of the drug development process.
Provided by American Chemical Society