Quantum biology and Ockham's razor
(PhysOrg.com) -- In a paper just published in Nature Chemistry, a team of University of Bristol scientists explores whether new models or concepts are needed to tackle one of the 'grand challenges' of chemical biology: understanding enzyme catalysis.
On a microscopic scale, nearly all the machinery that keeps our cells working involves chemical reactions of some sort and these reactions wouldn’t happen without enzymes – natural catalysts which make reactions happen very quickly, fast enough for life to be possible.
How do they do this? Understanding enzyme catalysis is a fundamental problem in biology. Billions of years of evolution have made enzymes superb catalysts, but the underlying physical principles are hotly debated. If we could design catalysts like them, they could transform areas such as ‘green’ energy generation and would make it possible to make new molecules in environmentally friendly ways. Engineered enzymes and designed protein catalysts are far less efficient than their natural counterparts, however. Chemists have yet to achieve the dream of making catalysts as powerful as natural enzymes.
This has led many researchers to propose that current theoretical models and concepts may not be adequate for understanding enzymes, and that new paradigms are required. This is particularly true for enzyme reactions that involve a particular quantum mechanical phenomenon known as tunnelling.
In quantum tunnelling, a particle passes through a barrier that, according to the laws of classical mechanics, it should not be able to get over. Scientists have known for some time that a whole host of reactions across chemistry and biology involve quantum tunnelling (in particular, the many reactions that involve transfer of particles as small as hydrogen atoms) and they have developed accurate models to describe its effects for chemical reactions.
However, some experimental observations for enzymes have led to recent suggestions that quantum tunnelling models for chemical reactions are inadequate when enzymes are involved. These ideas propose that enzymes somehow force tunnelling to happen in some special way – and that we therefore require new models for understanding how enzymes work. So: do we have the theoretical tools to understand enzymes, or are new models and concepts needed?
In a paper published in Nature Chemistry, Dr David Glowacki, Professor Jeremy Harvey and Professor Adrian Mulholland of Bristol’s School of Chemistry took Ockham’s razor to the enzyme tunnelling problem.
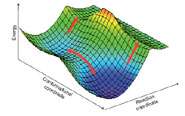
They used some simple maths to show that standard models for describing quantum tunnelling can explain the experimental enzyme data, so long as one accounts for the fact that enzymes have many possible different structures. Enzymes, like all proteins, are constantly fluctuating between many subtly different structures, which can have significantly different catalytic properties.
This dynamic nature of proteins has also been highlighted by scientists who study protein folding (the process by which a protein structure assumes its functional shape or conformation, the other ‘grand challenge’ of chemical biology). The Bristol researchers' paper indicates that enzyme structural fluctuations are significant, and by including them in modelling, scientists have the theoretical tools needed to understand these amazing natural catalysts. This should make it possible in future to design new catalysts using these principles.
More information: ‘Taking Ockham’s razor to enzyme dynamics and catalysis’ by David R. Glowacki, Jeremy N. Harvey and Adrian J. Mulholland in Nature Chemistry
Provided by University of Bristol












.jpg)