How Arctic microbes respond to a warming world

From the North Pole to the Arctic Ocean, the frozen soils within this region keep an estimated 1,672 billion metric tons of carbon out of the Earth's atmosphere. This sequestered carbon is more than 250 times the amount of greenhouse gas emissions attributed to the United States in the year 2009. As global temperatures slowly rise, however, so too do concerns regarding the potential impacts upon the carbon cycle when the permafrost thaws and releases the carbon that has been trapped for eons. Like so many of the planet's critical environmental processes, the smallest players—microbes—have the most significant influence over the eventual outcome.
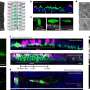
To answer this question, researchers from the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), the Earth Sciences Division (ESD) within Berkeley Lab, and the U.S. Geological Survey collaborated to understand how the microbes found in permafrost respond to their warming environment. Among the findings, published online November 6 in the journal Nature, is the draft genome of a novel microbe that produces methane, a far more potent greenhouse gas than carbon dioxide. This microbe, not yet named, lives in the permafrost, and was assembled out of the collection of genomes—the metagenome—isolated from the frigid soil. The assembly challenge is similar to building one complete jigsaw puzzle from a large collection of pieces from many different puzzles.
"The permafrost is poised to become a major source of greenhouse gases as the temperature in the Arctic is expected to increase dramatically compared to the expected temperature increase in many other regions of the world," said ESD's Janet Jansson, corresponding author and initiator of the study (first supported by a grant to her from DOE Laboratory Directed Research and Development funds). "By applying metagenomics to study microbial community composition and function, we can help to answer questions about how the currently uncultivated and unstudied microbial species residing in permafrost cycle organic carbon and release greenhouse gases during thaw," Jansson said. "This will provide valuable information that could lead to improved carbon cycle models and eventual mitigation strategies."
According to the U.S. Environmental Protection Agency, in 2009 fossil fuel combustion accounted for 5.2 billion metric tons of the nation's carbon dioxide emissions, a tiny fraction (about 3/10 of 1 percent) of the carbon dioxide trapped in the Arctic permafrost. Understanding the microbial processes in the frozen soils and the impacts of microbial activity on carbon dioxide processes, has been a project of study co-author and USGS researcher Mark Waldrop, whose 2010 Community Sequencing Program project, another with the DOE JGI (in collaboration with Jansson), focuses on comparing the short-term microbial response of the thawing permafrost to the longer-term processes.
Waldrop cored meter-deep permafrost samples from a peaty black spruce forest along Hess Creek, Alaska. Each sample was then divided to distinguish between the seasonally thawed "active" layer, which comprised the top third of the core, and the permanently frozen "permafrost" horizon underneath. The samples were incubated during thaw at 5°C and then sent to the Jansson lab for DNA extraction and analyses At the DOE JGI, the team led by Director Eddy Rubin and his postdoctoral fellow Rachel Mackelprang, (now at California State University-Northridge) sequenced microbial DNA from the samples. "These microbial communities are extremely diverse," said Mackelprang. "A single gram of soil may contain thousands of different bacterial species and billions of cells. Additionally, most of these microbes cannot be grown in the laboratory, making this an extremely difficult area to study." The data were then analyzed in collaboration with Jansson and her team at ESD, including Kristen DeAngelis, Maude David, and Krystle Chavarria.
At DOE JGI, Mackelprang and her colleagues generated nearly 40 billion bases of raw DNA sequence, necessary due to the high microbial diversity of the soil. They identified several microbes that produced methane as a byproduct, and were able to assemble a draft genome of a novel methanogen. "This is the first example of a successful assembly of a draft genome from a highly complex soil metagenome," the authors wrote. "The abundance of this novel methanogen suggests that it may be an important player in methane production under frozen conditions." Additionally, the genomic data revealed that the microbe had genes for nitrogen fixation, making this study also the first to describe a potentially nitrogen-fixing methanogen in permafrost soil.
The researchers identified many genes involved in carbon and nitrogen cycling in the metagenomic data, and found that their levels of abundance shifted in response to their thawing habitat. "These detailed analyses reveal for the first time the rapid and dynamic response of permafrost microbial communities to thaw," they concluded. "The thaw-induced shifts that we detected directly support conceptual models of carbon and nitrogen cycling in arctic soils, in which microbes play a central role in greenhouse gas emissions and destabilization of stored permafrost carbon."
Provided by DOE/Joint Genome Institute