A hot species for cool structures: Complex proteins in 3-D thanks to simple heat-loving fungus
A fungus that lives at extremely high temperatures could help understand structures within our own cells. Scientists at the European Molecular Biology Laboratory (EMBL) and Heidelberg University, both in Heidelberg, Germany, were the first to sequence and analyse the genome of a heat-loving fungus, and used that information to determine the long sought 3-dimensional structure of the inner ring of the nuclear pore. The study was published today in Cell.
The fungus Chaetomium thermophilum lives in soil, dung and compost heaps, at temperatures up to 60 C. This means its proteins – including some which are very similar to our own – have to be very stable, and the Heidelberg scientists saw this stability as an advantage.
"There are a number of structures that we couldn't study before, because they are too unstable in organisms that live at more moderate temperatures," explains Peer Bork, who led the genome analysis at EMBL. "Now with this heat-loving fungus, we can."
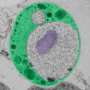
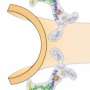
The scientists compared the fungus' genome and proteome to those of other eukaryotes – organisms whose cells have a nucleus – and identified the proteins that make up the innermost ring of the nuclear pore, a channel that controls what enters and exits a cell's nucleus. Having identified the relevant building blocks, the scientists determined the complex 3D structure of that inner ring for the first time.
"This work shows the power of interdisciplinary collaborations," says Ed Hurt, who led the structural and biochemical analyses at Heidelberg University: "the nuclear pore is an intricate biological puzzle, but by combining bioinformatics with biochemistry and structural biology, we were able to solve this piece of it for the first time."
The scientists have made C. thermophilum's genome and proteome publicly available, and are confident that these will prove valuable for studying other eukaryotic structures and their interactions, as well as general adaptations to life in hot places. Such knowledge could potentially lead to new biotechnology applications.
More information: Published online in Cell on 22 July 2011.
Provided by European Molecular Biology Laboratory