Non-independent mutations present new path to evolutionary success
Mutations of DNA that lead to one base being replaced by another don't have to happen as single, independent events in humans and other eukaryotes, a group of Indiana University Bloomington biologists has learned after surveying several creatures' genomes.
And, the scientists argue, if "point mutations" can happen in twos, threes -- even nines – large evolutionary jumps are possible, especially when problems caused by a single point mutation are immediately compensated for by a second or third. The work appears in the latest issue of Current Biology.
"A similar phenomenon had been observed in bacteria," said Matthew Hahn, the project's principal investigator. "And the idea that this might be happening in eukaryotes has been around for a while. We are the first ones to use exhaustive genomic studies to show it's actually happening, and happening in a big way."
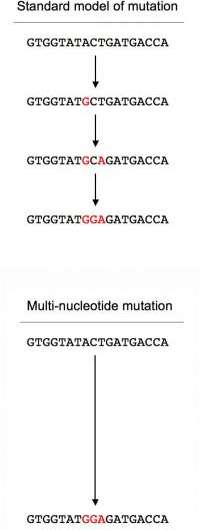
Hahn and two members of his lab, Ph.D. student Daniel Schrider and undergraduate Jonathan Hourmozdi, surveyed the disparate genomes of yeast (Saccharomyces cerevisiae), roundworm (Caenorhabditis elegans), fruit fly (Drosophila melanogaster), the model plant Arabidopsis thaliania and humans, and found that across the board, about three percent of new mutations are "multi-nucleotide mutations," or MNMs, perhaps the result of a single, error-prone DNA polymerase making two or more mistakes as it made its way down the chromosome. The group also studied human trios of parent-parent-offspring DNA, as well as the complete genomes of a Yan Chinese (YH01) and J. Craig Venter, cofounder of (and donor to) the Human Genome Project. The researchers found tens of thousands of likely MNMs.
MNMs were essentially defined by the proximity of two or more point mutations. Since mutations are rare, the statistical likelihood of finding two mutations within 20 or 100 bases of each other after a few generations (or a few replication events in the germ line) is low enough to assume two nearby mutations have a near-100 percent likelihood of being caused by the same mutational event.
Three percent of new mutations may not sound like a lot, but even rare genetic phenomena can be very powerful if they impact a creature's fitness, the measure of an individual's ability to survive and reproduce.
"There are cases where an organism could improve its fitness if it acquired multiple mutations that would each reduce fitness if they occurred individually," Schrider said. "In cases like this, the organism would not be able to reach the improved fitness state, as the less-fit intermediate states would be eliminated by natural selection. Cases like this are referred to as 'fitness valleys.'"
The exchange of a single base within a gene can have drastic consequences for the behavior of the protein that gene encodes. Sickle cell anemia, for example, which causes red blood cells to become rigid, sharp-edged, and resistant to oxygen absorption -- and is the cause of listlessness and excruciating pain in the humans who have it -- is caused by a point mutation.
The idea, Hahn and Schrider say, is that whatever problems a point mutation causes could be ameliorated by a second, with one point mutation compensating for the other in between generations. The scientists admit they expect this would be a very rare event. But possible.
"The most exciting implication of our work is that it raises the possibility that organisms could leap across fitness valleys and reach a higher-fitness state by acquiring multiple mutations simultaneously," Schrider said.
Hahn says he does not yet know of examples of genes in which valley leaping might have occurred, but that he and other researchers are eager to investigate.
"Our work provides evidence for a possible new mechanism of adaptation," Hahn said. "It also raises questions about whether thousands of supposedly independent mutations others have observed in important genes are truly independent. Some of these genes will need to be reanalyzed, because the recognition that some of these mutations are actually MNMs could have an impact on many analyses of DNA sequences."
But first, Hahn's group will try to see whether the MNMs they've found in humans conform to the mechanism geneticists have observed in humans' faultier DNA polymerases.
"It's satisfying to be able to be able to show that this is real," Hahn said. "I talked about this at a recent conference, and no matter who we talked to, they said the same sorts of things: 'Oh yeah I've seen those before, but I just thought it was a statistical anomaly.' People have seen the phenomenon but they just didn't know it was meaningful. We hope this gets people excited to go back and look at their old sequence data."
Provided by Indiana University