New protein identified in bacterial arsenal
(PhysOrg.com) -- Nearly a billion years ago, bacteria evolved an insidious means of infecting their hosts — a syringe-like mechanism able to inject cells with stealthy hijacker molecules. These molecules, called virulence factors, play a sophisticated game of mimicry, imitating many of the cells’ normal activities but ultimately co-opting them to serve the bacteria’s needs. Now researchers at The Rockefeller University have identified a new class of these coup artists that appear to take over a key process that regulates a wide range of cellular duties, from cell-cycle progression to cell death, even communication between cells.
Scientists in C. Erec Stebbins’s Laboratory of Structural Microbiology have discovered the crystal structure of virulence factor SspH2, which is deployed by Salmonella, a mean strain of bacteria that can cause food poisoning, typhoid fever and septicemia. With colleagues at Yale University, they performed a series of biochemical experiments to show that SspH2 is an enzyme that links two molecules together, called a ligase. Specifically, it is involved in the targeting of a widespread regulatory molecule called ubiquitin to other proteins. Ubiquitin’s most common function is to label proteins for degradation in a process called ubiquitination.
“It’s a totally new ubiquitin ligase from a bacterial pathogen going in and messing around with human cell chemistry,” says Stebbins. “It’s a good example of the amazing ways bacteria have found to play around with our biochemistry for its own purposes.”
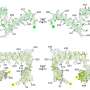
The structure, derived from the pattern of light bouncing off of a crystallized form of SspH2, reveals the identity and position of every atom in the virulence factor. It shows that it has two primary arms: One is a known structure that recruits other proteins, but the second had never been seen before. The Stebbins team named it NEL for Novel E3 Ligase. The two arms conceal a key amino acid — cysteine — required to bind to ubiquitin. In test-tube experiments, Stebbins and colleagues showed that in order to capture the ubiquitin, one arm must swing apart to expose the cysteine, and moreover, it must do so in a selective fashion. They generated a version of the molecule effectively locked in its open state and found that it was toxic to the cells it invaded.
The process of attaching ubiquitin to targeted proteins — ubiquitination — is very common in the cells of complex organisms and involves three different types of molecules such as SspH2. Molecules known as E1 capture the ubiquitin and hand it off to E2 molecules. E3 molecules then bring the E2 and its ubiquitin to attach to specific proteins. Stebbins and colleagues determined that SspH2 is a new member of this third family and that it targets proteins on top of cell membranes, but exactly which ones remain to be seen.
“When we find that out, we’ll know exactly what this ubiquitin ligase is doing,” Stebbins says. The findings are published this week in the Proceedings of the National Academy of Sciences.
More information: Proceedings of the National Academy of Sciences online, A family of Salmonella virulence factors functions as a distinct class of E3 ubiquitin ligases, Cindy M. Quezada, Stuart W. Hicks, Jorge E. Galán and C. Erec Stebbins
Provided by Rockefeller University