This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
Decoding Atractylodes lancea: A genomic journey through adaptation and metabolism
The cultivation of high-quality medicinal plants like Atractylodes lancea involves intricate genetic processes influenced by environmental factors. Despite its extensive use in traditional medicine, the genetic basis and metabolomic diversity of A. lancea remain largely unexplored.
Growing in varied environments, the plant encounters challenges such as drought and nutrient deficiencies, driving genetic differentiation that impacts its medicinal qualities. Understanding these genetic mechanisms is essential for enhancing the quality and adaptability of A. lancea under changing environmental conditions.
Researchers from the State Key Laboratory for Quality Ensurance and Sustainable Use of Dao-di Herbs, China Academy of Chinese Medical Sciences, have published a study in Horticulture Research, unraveling the genetic and metabolomic foundations of population evolution and local adaptation in Atractylodes lancea.
By assembling a high-quality genome and examining population resequencing data, the team identified crucial genetic variations influencing the plant's metabolome across regions. These findings shed light on the genetic regulation of A. lancea's medicinal traits.
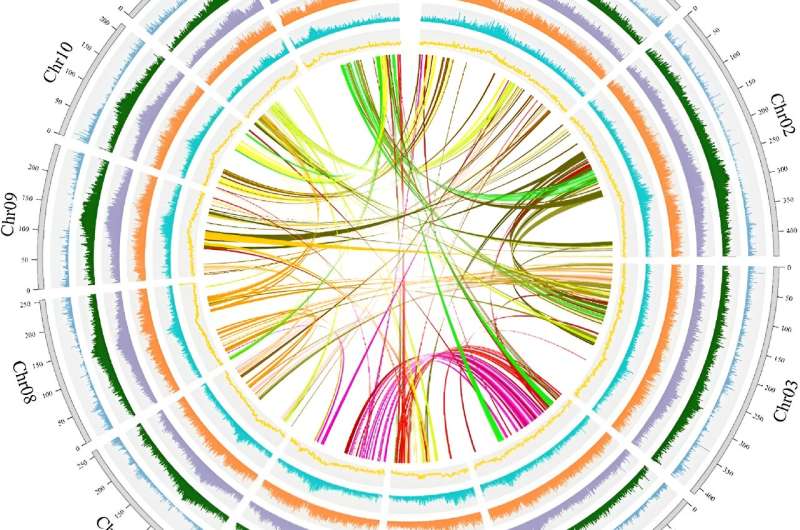
The study assembled a high-quality, chromosome-scale genome of Atractylodes lancea, revealing a genome size of about 4,009 Mb with a contig N50 of 1.18 Mb. Population resequencing of 251 samples from 22 locations revealed significant genetic divergence among three major groups: Maoshan-Dabie Mountains (MA), North Yanshan Mountain (NA), and Qinling-Taihang Mountains (SA).
Each group exhibited distinct metabolomic profiles, particularly in key medicinal compounds like β-eudesmol, hinesol, and atractylodin. A genome-wide association study (GWAS) identified genes such as AlZFP706 and AlAAHY1, which are linked to the accumulation of these bioactive compounds.
The study highlights how natural selection, including environmental pressures like drought and potassium deficiency, shapes the genetic and metabolic diversity of A. lancea, driving its local adaptation. This comprehensive dataset offers valuable resources for the genetic improvement of A. lancea and its medicinal applications.
"Deciphering the genetic basis of local adaptation in medicinal plants like Atractylodes lancea is vital for enhancing their quality and sustainability," said Dr. Sheng Wang, lead author of the study.
"Our research offers a detailed view of how natural variations impact the genetic and metabolic landscape of A. lancea, providing insights into the evolutionary mechanisms behind its medicinal properties. This knowledge not only expands our understanding of plant adaptation but also supports the development of better cultivation strategies to maintain the herb's therapeutic quality."
The study's findings have significant implications for the cultivation and quality control of Atractylodes lancea. Identifying key genetic markers associated with medicinal traits allows for targeted breeding strategies to enhance desirable qualities, ensuring the herb's effectiveness as a traditional remedy.
Moreover, understanding the genetic and metabolic adaptations of A. lancea can support the verification of its geographic origin, maintaining the authenticity of medicinal products. This research paves the way for the genetic enhancement and sustainable use of traditional medicinal plants.
More information: Chengcai Zhang et al, Genome resequencing reveals the genetic basis of population evolution, local adaptation, and rewiring of the rhizome metabolome in Atractylodes lancea, Horticulture Research (2024). DOI: 10.1093/hr/uhae167
Journal information: Horticulture Research
Provided by TranSpread