This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Exploring the molecular basis of how pradimicin A binds to viral N-glycan, a potential SARS-CoV-2 entry inhibitor
HIV, Ebola and most recently, COVID-19 viruses have had an enormous impact on our societies world-wide. All these viruses are "enveloped viruses," viruses that have an exterior envelope that surrounds them largely composed of their host's cells. This envelope increases the virus's ability to hide from its host's immune system and to access the host's cells. It also, however, gives researchers a target, an opportunity to interrupt viral transmission.
Japanese researchers have been working on the issue of halting viral transmission in these types of viruses.
"The development of vaccines and antiviral drugs against COVID-19 has successfully reduced the risk of death, but complete suppression of viral transmission is still challenging. Under such circumstances, we evaluated the potential of naturally occurring pradimicin A (PRM-A) as a new anti-SARS-CoV-2 drug that suppresses SARS-CoV-2 transmission," said Yu Nakagawa, the lead author on the paper and an associate professor in the Institute for Glyco-core Research (iGCORE) at Nagoya University, Nagoya, Japan.
Their research was published in Bioorganic & Medicinal Chemistry on May 1.
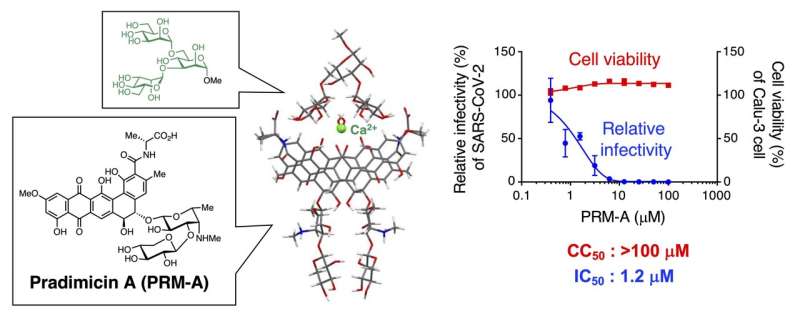
There is strong evidence that PRM-A is a viral entry inhibitor; in other words, it stops viruses from entering a host's cells. It does this by binding to N-glycans, which are found on the surface of several types of enveloped viruses including the SARS-CoV-2 virus. However, there is still little known about how exactly PRM-A binds to the viral N-glycans.
To infect a cell, a virus's envelope uses specific receptors on its surface called spike proteins—which are usually glycoproteins, meaning carbohydrates, specifically sugar (oligosaccharides) attached to proteins—to bind to the cellular membrane of a host's cell, causing a conformational change in the cell membrane which allows the virus to enter the cells. Once there, it uses the cell's resources to replicate its own genome, safe from the host's immune system.
Initially, researchers looking at interrupting viral transmission focused on lectins, carbohydrate-binding proteins that are derived from plants or bacteria, which showed strong promise as a viral entry inhibitor. They bind with the viruses' glycoproteins and stop its advance into a cell.
However, they are often expensive, easily targeted by the host's immune system, and may be toxic to the host's cells. Lectin mimics have many of the carbohydrate-binding ability of the lectin without the expensive and dangerous side effects.
The Japanese team looked at PRM-A, a naturally occurring lectin mimic. It has shown promise as a viral entry inhibitor as there is evidence it binds to the N-glycans of the viruses' envelope glycoproteins.
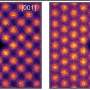
To determine the molecular basis of the binding, they used molecular modeling and ran binding assays which measure the reactions between PRM-A and N-glycans as they bind. They also carried out in vitro experiments to test PRM-A's ability to inhibit SARS-CoV-2.
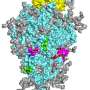
They found that PRM-A binds selectively to branched oligomannose structures found in high mannose-type and hybrid-type N-glycans on viral spike proteins. Mannose is the specific sugar found in these N-glycans. They also found that PRM-A did inhibit the infectivity of SARS-CoV-2. In fact, the inhibition occurred through the interaction between the PRM-A and the branched oligomannose-containing N-glycans.
"We demonstrated for the first time that PRM-A can inhibit SARS-CoV-2 infection by binding to viral glycans. It is also noteworthy that PRM-A was found to bind preferentially to branched oligomannose motifs of viral glycans via simultaneous recognition of two terminal mannose residues. This finding provides essential information needed to understand the antiviral mechanism of PRM-A," said Nakagawa.
Nakagawa and their team are already busy working on the next step in their research.
"Our ultimate goal is to develop anti-SARS-CoV-2 drugs based on PRM-A. The glycan-targeted antiviral action of PRM-A has never been observed in major classes of the existing chemotherapeutics, underscoring its potential as a promising lead for antiviral drugs with the novel mode of action.
"Especially, considering that glycan structures are hardly changed by viral mutation, we expect that PRM-A-based antiviral drugs would be effective against mutated viruses. Toward this goal, we are now examining in vivo antiviral activity of PRM-A using hamsters, and also developing PRM-A derivatives that are more suitable for therapeutic applications," said Nakagawa.
More information: Yu Nakagawa et al, Molecular basis of N-glycan recognition by pradimicin a and its potential as a SARS-CoV-2 entry inhibitor, Bioorganic & Medicinal Chemistry (2024). DOI: 10.1016/j.bmc.2024.117732
Provided by Institute for Glyco-core Research (iGCORE), Tokai National Higher Education and Research System