This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
Unlocking the genetic secrets of cabbage: Key inversions suppress recombination for crop improvement
Chromosomal inversions are structural variations that play a significant role in suppressing genetic recombination, thereby fixing favorable allelic combinations. These inversions are crucial for maintaining coadapted genotypes and are involved in species evolution.
In rice and tomato, similar inversions have been identified, highlighting their importance in plant genetics. Due to these challenges, it is essential to conduct in-depth research on chromosomal inversions to enhance our understanding of their mechanisms and applications in crop improvement.
Based on these challenges, a detailed study on chromosomal inversions in cabbage was necessary.
A recent study conducted by the State Key Laboratory of Vegetable Biobreeding at the Chinese Academy of Agricultural Sciences and the State Key Laboratory of Crop Genetics and Germplasm Enhancement at Nanjing Agricultural University was published in the journal Horticulture Research on January 30, 2024.
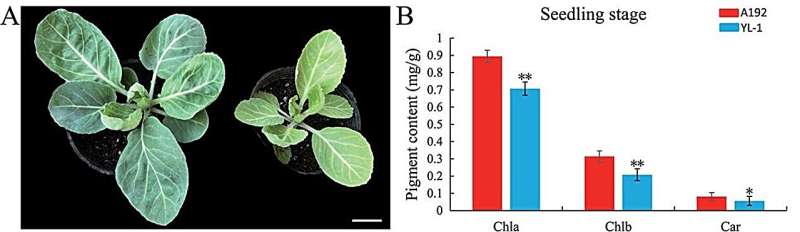
This research, focusing on the yellow–green leaf mutant YL-1 in cabbage, has identified two large chromosomal inversions that significantly suppress recombination.
The study investigates the genetic mechanisms of the yellow–green leaf mutant YL-1 in cabbage, identifying two significant chromosomal inversions, INV1 and INV2, on chromosome 1 within the BoYgl-1 locus. These inversions cause severe recombination suppression, crucial for fixing key genotypes in cabbage breeding.
Chlorophyll and carotenoid contents were significantly reduced in YL-1 compared to the normal-green leaf A192, impacting photosynthetic efficiency. Using advanced genomic technologies, including Illumina, PacBio, and Hi-C sequencing, the YL-1 genome was assembled and compared with the TO1000 reference genome.
These inversions, found in 44 out of 195 cabbage lines, highlight their evolutionary significance.
Dr. Fengqing Han, a leading scientist at the Chinese Academy of Agricultural Sciences, stated, "The discovery of these large chromosomal inversions opens up new possibilities for genetic research and breeding in cabbage. Understanding the mechanisms of recombination suppression will enable us to develop more resilient and commercially valuable cabbage varieties."
The research offers tools for breeders to develop cabbage varieties with better disease resistance, nutritional content, and climate adaptability. Understanding recombination suppression can create stable genotypes with beneficial traits, paving the way for similar studies in other crops, transforming agriculture and food security.
More information: Bin Zhang et al, Two large inversions seriously suppress recombination and are essential for key genotype fixation in cabbage (Brassica oleracea L. var. capitata), Horticulture Research (2024). DOI: 10.1093/hr/uhae030
Journal information: Horticulture Research
Provided by TranSpread