This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
New simplified DNA model for advanced computational simulations
DNA is the molecule that contains all the genetic information necessary for the development and functioning of living organisms. It is organized in a structure called "chromatin," which is found inside the nucleus of cells. The shape that chromatin takes directly affects the activity of genes, and it is therefore important to have a detailed understanding of the structure of DNA and to be able to predict its variations.
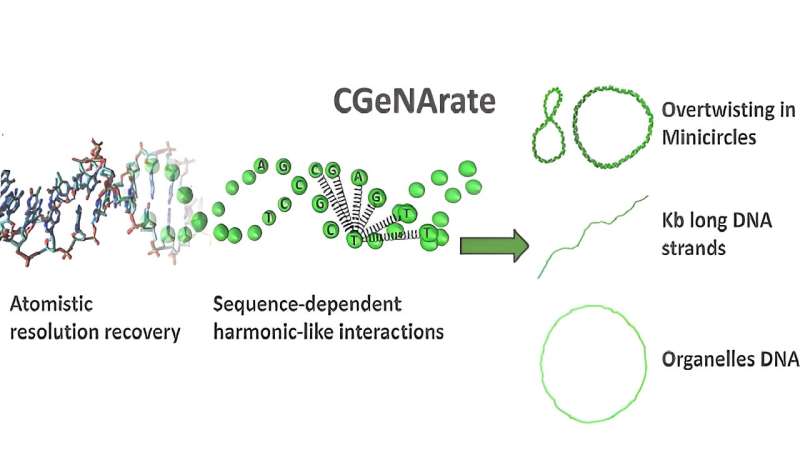
Now, a paper published in the journal Nucleic Acids Research presents a simplified DNA model called CGeNArate, which allows fast and accurate computational simulations to be carried out.
The study was led by Modesto Orozco, professor at the Faculty of Chemistry of the University of Barcelona and head of the Molecular Modeling and Bioinformatics Laboratory at the Institute for Research in Biomedicine (IRB Barcelona), located at the Barcelona Science Park (PCB).
The method uses a specific approach that consists of simplifying a complex system by reducing its number of degrees of freedom and grouping atoms into coarse-graining units. In addition, it uses machine learning algorithms to reconstruct the atomic resolution of the simulated trajectories. This allows the structure and behavior of DNA to be studied without the need for complex and time-consuming models, facilitating faster and more accessible research.
"This new model can be useful for projects that require studying the DNA's structure in a detailed and agile way. Although the model is already functional, we are working to improve it and apply it into other components of the cell nucleus, which could open new opportunities in biomedical research," notes Modesto Orozco, professor at the UB's Department of Biochemistry and Molecular Biomedicine.
Detailed genomic information
The research team tested the new tool and validated its potential for obtaining high-speed chromatin simulations. The model allowed the observation of the behavior of a specific section of DNA in a yeast gene, as well as modeling the structure of DNA in mitochondria and reproducing the flexibility of different DNA structures in various contexts.
"It was also able to simulate the formation of small circular DNA structures known as minicircles," says David Farré-Gil, first author of the research and a Ph.D. student in the same lab. These results demonstrate the effectiveness of the method for detailed analysis of DNA and chromatin.
More information: David Farré-Gil et al, CGeNArate: a sequence-dependent coarse-grained model of DNA for accurate atomistic MD simulations of kb-long duplexes, Nucleic Acids Research (2024). DOI: 10.1093/nar/gkae444
Journal information: Nucleic Acids Research
Provided by University of Barcelona