This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Team develops efficient host-vector system for a model archaeon by solving CRISPR-based host-plasmid conflict
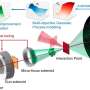
A research group has constructed versatile genetic tools for Saccharolobus islandicus REY15A, one of the very few archaeal models for archaea biology and CRISPR biology research.
Such tools include efficient genome editing, robust protein expression systems, interference plasmid assay, gene silencing and CRISPR-based gene editing. Nevertheless, plasmid vectors constructed for this crenarchaeon thus far are based solely on the pRN2 cryptic plasmid.
The study, appearing in mLife, was led by Prof. Qunxin She and Dr. Guanhua Yuan, both of Shandong University in Qingdao, China.
"A dual host-vector system is required to enrich the genetic toolbox for this model archaeon," says Prof. She.
In fact, at an early stage of archaeal vector development, both pRN1 and pRN2 plasmids that coexist in the Sa. islandicus REN1H1 were employed for constructing shuttle vectors for Sa. islandicus REY15A based on these two plasmids; pRN2‐derived plasmids scored a high transformation rate and yielded true transformants, while pRN1‐based vectors yielded only very few colonies from which plasmids were apparently absent.
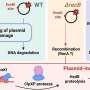
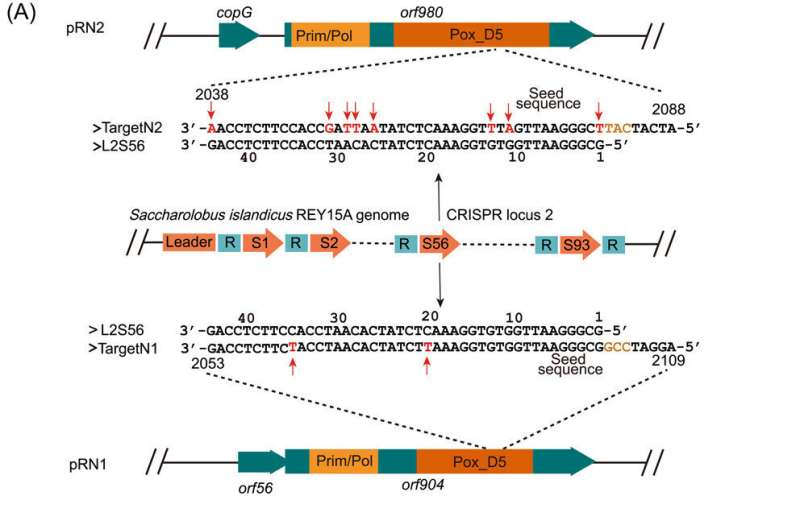
"Since CRISPR arrays often carry spacers matching various plasmids in Sulfolobales, we suspected that the genome of Sa. islandicus REY15A might carry a spacer that matches a sequence in pRN1 but not in pRN2," Dr. Yuan says.
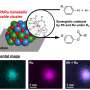
After determining the complete genome sequence of Sa. islandicus REY15A4, the host genome does carry a spacer (L2S56) only showing two mismatches to a DNA segment (Target N1) in the coding sequence of the pRN1 replicase gene. Transformation efficiency experiments demonstrated that L2S56 crRNAs were expressed to a level that could be sufficient to elicit the I‐A immunity but insufficient to trigger the III‐B immunity for plasmid elimination in Sa. islandicus REY15A.
To obtain a functional target N1 derivative that evades the host CRISPR immunity, the team designed three DNA segments (N1a, N1b and N1c) based on the pRN1 target, while the designed mutations in N1a were synonymous, N1b and N1c had missense mutations. The results showed that none of the three mutated targets were targeted by the CRISPR system in the archaeal host. However, subsequent experiments showed that N1c carries the missense mutations that may have inactivated the replication protein.
Through the construction of a series of vectors, the Saccharolobus–E. coli shuttle vector pN1dAA with the N1a mutations, argD selection marker, p15A origin of replication and a kanamycin‐resistant marker were designed based on the pRN1 backbone, which can achieve a stable coexistence with the pRN2-derived plasmid pSeSD in Sa. islandicus REY15A cells. This yielded a dual plasmid system for genetic study with this important archaeal model.
Since examination of host-plasmid conflicts provides a useful means for identification of compatible plasmid-vector systems, once the conflict is solved experimentally, the engineered plasmids are then very useful for developing host-vector systems, as reported in this article.
More information: Pengpeng Zhao et al, Rational design of unrestricted pRN1 derivatives and their application in the construction of a dual plasmid vector system for Saccharolobus islandicus, mLife (2024). DOI: 10.1002/mlf2.12107
Provided by Tsinghua University Press