This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Research team develops new technique to release and study individual proteins in cells
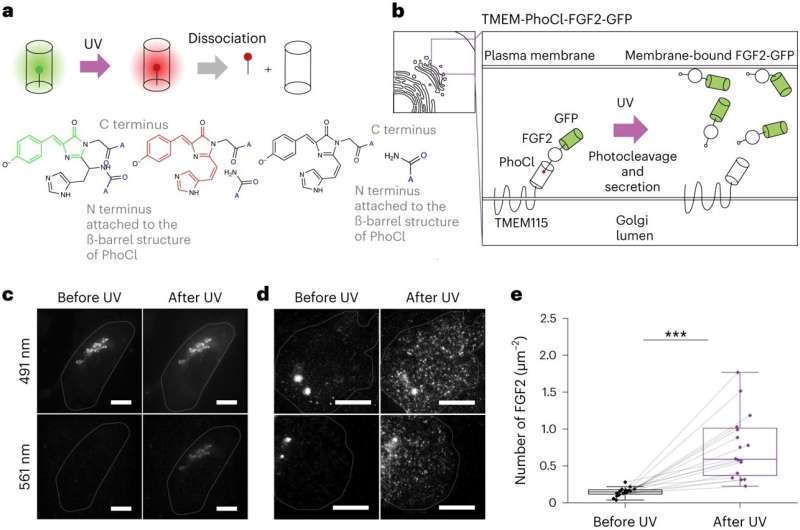
A research team led by biochemist Professor Helge Ewers from Freie Universität Berlin has developed a new technique for the light-mediated release and investigation of proteins in live cells. The technique makes use of a laser pulse to control the release of tagged protein molecules within a cell, allowing for the molecules' function to be more clearly observed. The team believes that this method has a wide variety of potential applications in future scientific research.
At the simplest level, proteins can be understood as tiny machines—a mere millionth of a millimeter in size—that are responsible for carrying out the majority of functions in our cells. In an attempt to better understand these proteins, the Ewers Group at Freie Universität Berlin's Institute of Chemistry and Biochemistry has developed a method through which to observe cellular function at the molecular level better.
Single molecule microscopy, i.e., the ability to observe single molecules, has greatly added to our understanding of certain mechanisms that take place within cells, for example, how genetic material is copied. However, one issue with this technology to date is that each cell contains thousands of copies of each protein. This makes it difficult to follow proteins individually to investigate their function within the context of the cell, even when they have been tagged with color.
Ewers and his research team have now developed a new technique that allows scientists to observe a select number of tagged proteins at the same time. The technique works by binding tagged proteins to the Golgi apparatus—an organelle found in all plant and animal cells—using a special protein. A short, controlled pulse of light then severs this connection, allowing for some of the proteins to be transported to the place in the cell where they carry out their function.
"This new technique represents a breakthrough in studying individual proteins in cells. With just a short pulse of light, we can release single molecules in cells in a controlled manner and, in doing so, better observe their function," explains Ewers, who leads the research group at Freie Universität.
Dr. Purba Kashyap, cell biologist at Freie Universität Berlin and first author of the study, emphasizes, "Once we saw that we could control the number of proteins that were released by varying the intensity of the laser beam, we knew that our technique could have a wide variety of applications and be of great help to other researchers."
Details of the technique, which was developed by the working group at Freie Universität together with other laboratories in Berlin, Hamburg, and Tokyo, have been published in the latest edition of Nature Methods.
"This method is now well established in three different laboratories. It has proven to be very robust, and we are already in touch with several parties who are interested in its scientific applications," says Ewers. Potential future areas of application include manipulating protein functions in infected cells or in the individual cells of living organisms, not to mention the general improvements this technique represents for research that requires directly observing individual proteins and their functions.
"For example, the ability to manipulate exactly how many proteins are released will enable us to count exactly how many mutated receptors are required to make a cell reproduce itself uncontrollably, for example, in diseases such as cancer."
Ewers and his team are already making plans to implement this technique with external partners, as well as to develop it further, "Berlin is an important hub for optogenetics research—a field that explores how light can be used to solve biological problems. And we are delighted that we were able to collaborate so efficiently with Professor Andrew Plested (Humboldt-Universität zu Berlin) and Dr. Marcus Taylor (Max Planck Institute for Infection Biology)."
Together, the working groups were able to demonstrate how the new technique could be used to restore protein functionality in immune cells with genetic defects. Collaborations with other working groups will be aimed at putting the new method into practice in fruit flies in the near future.
More information: Purba Kashyap et al, An optogenetic method for the controlled release of single molecules, Nature Methods (2024). DOI: 10.1038/s41592-024-02204-x
Provided by Free University of Berlin