This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
New software allows scientists to model shapeshifting proteins in native cellular environments
Cells rely on complex molecular machines composed of protein assemblies to perform essential functions such as energy production, gene expression, and protein synthesis. To better understand how these machines work, scientists capture snapshots of them by isolating proteins from cells and using various methods to determine their structures. However, this process also removes them from the context of their native environment, including protein interaction partners and cellular location.
Recently, cryogenic electron tomography (cryo-ET) has emerged as a way to observe proteins in their native environment by imaging frozen cells at different angles to obtain three-dimensional structural information. This approach is exciting because it allows researchers to directly observe how and where proteins associate with each other, revealing the cellular neighborhood of those interactions within the cell.
With the technology available to image proteins in their native environment, MIT graduate student Barrett Powell wondered if he could take it one step further: What if molecular machines could be observed in action? In a paper published March 8 in Nature Methods, Powell describes the method he developed, called tomoDRGN, for modeling structural differences of proteins in cryo-ET data that arise from protein motions or proteins binding to different interaction partners. These variations are known as structural heterogeneity.
Although Powell had joined the lab of MIT associate professor of biology Joey Davis as an experimental scientist, he recognized the potential impact of computational approaches in understanding structural heterogeneity within a cell. Previously, the Davis Lab developed a related methodology named cryoDRGN to understand structural heterogeneity in purified samples. As Powell and Davis saw cryo-ET rising in prominence in the field, Powell took on the challenge of re-imagining this framework to work in cells.
When solving structures with purified samples, each particle is imaged only once. By contrast, cryo-ET data is collected by imaging each particle more than 40 times from different angles. That meant tomoDRGN needed to be able to merge the information from more than 40 images, which was where the project hit a roadblock: The amount of data led to an information overload.
To address this, Powell successfully rebuilt the cryoDRGN model to prioritize only the highest-quality data. When imaging the same particle multiple times, radiation damage occurs. The images acquired earlier, therefore, tend to be of higher quality because the particles are less damaged.
"By excluding some of the lower-quality data, the results were actually better than using all of the data—and the computational performance was substantially faster," Powell says.
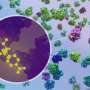
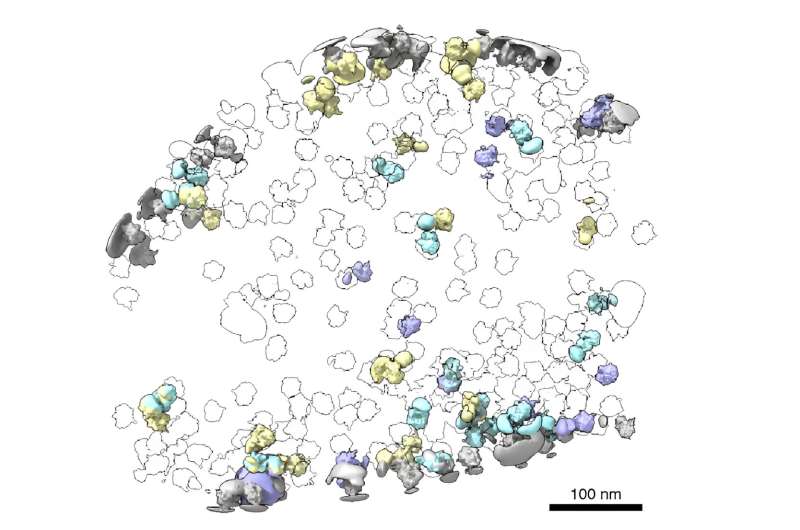
Just as Powell was beginning work on testing his model, he had a stroke of luck: The authors of a groundbreaking new study that visualized, for the first time, ribosomes inside cells at near-atomic resolution, shared their raw data on the Electric Microscopy Public Image Archive (EMPIAR). This dataset was an exemplary test case for Powell, through which he demonstrated that tomoDRGN could uncover structural heterogeneity within cryo-ET data.
According to Powell, one exciting result is what tomoDRGN found surrounding a subset of ribosomes in the EMPIAR dataset. Some of the ribosomal particles were associated with a bacterial cell membrane and engaged in a process called cotranslational translocation. This occurs when a protein is being simultaneously synthesized and transported across a membrane.
Researchers can use this result to make new hypotheses about how the ribosome functions with other protein machinery integral to transporting proteins outside of the cell, now guided by a structure of the complex in its native environment.
After seeing that tomoDRGN could resolve structural heterogeneity from a structurally diverse dataset, Powell was curious: How small of a population could tomoDRGN identify? For that test, he chose a protein named apoferritin, which is a commonly used benchmark for cryo-ET and is often treated as structurally homogeneous. Ferritin is a protein used for iron storage and is referred to as apoferritin when it lacks iron.
Surprisingly, in addition to the expected particles, tomoDRGN revealed a minor population of ferritin particles—with iron bound—making up just 2% of the dataset, that was not previously reported. This result further demonstrated tomoDRGN's ability to identify structural states that occur so infrequently that they would be averaged out of a 3D reconstruction.
Powell and other members of the Davis Lab are excited to see how tomoDRGN can be applied to further ribosomal studies and to other systems. Davis works on understanding how cells assemble, regulate, and degrade molecular machines, so the next steps include exploring ribosome biogenesis within cells in greater detail using this new tool.
"What are the possible states that we may be losing during purification?" Davis asks. "Perhaps more excitingly, we can look at how they localize within the cell and what partners and protein complexes they may be interacting with."
More information: Barrett M. Powell et al, Learning structural heterogeneity from cryo-electron sub-tomograms with tomoDRGN, Nature Methods (2024). DOI: 10.1038/s41592-024-02210-z
Provided by Massachusetts Institute of Technology
This story is republished courtesy of MIT News (web.mit.edu/newsoffice/), a popular site that covers news about MIT research, innovation and teaching.