This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
What makes a pathogen antibiotic-resistant?
Antimicrobial resistance is a story of constantly moving parts and players. With every new or tweaked antibiotic or antimicrobial drug, the targeted pathogens begin the evolutionary dance of acquiring resistance, prompting researchers to constantly develop workarounds or entirely new classes of medicine.
Understanding the underlying mechanisms of acquired antimicrobial resistance is critical to the fight, a case of knowing one's enemy. In a new paper published in npj Antimicrobials and Resistance, researchers at Sanford Burnham Prebys, working with Roche Pharma Research and Early Development, describe how two notable pathogens—Escherichia coli and Acinetobacter baumannii—employ distinctly different tools to fend off an antibiotic attack.
"This work was conceived as a comparative study of the mechanisms and dynamics of resistance acquisition for two drugs and two bugs," said senior author Andrei L. Osterman, Ph.D., at Sanford Burnham Prebys.
"Comparing mutational landscapes triggered by the same drug in two distinct bugs allows us to deduce both shared and unique evolutionary trajectories toward resistance. A comparison of two drugs in the same bug reveals shared and unique mechanistic features of fundamental and translational importance, from drug discovery to rational optimization of treatment regimens."
Both pathogens studied are gram-negative bacteria with shared characteristics but also notable differences.
For the most part, E. coli is a bacterium routinely found in the guts of humans and animals, where it resides with no ill effect. Some strains, however, do cause harm, everything from mild gastroenterological distress to urinary tract infections, respiratory illness, and pneumonia.
Acinetobacter baumannii is more problematic, particularly in clinical settings where it can cause severe infections, some life-threatening.
Both bacteria have developed resistance to most current antibiotic treatments.
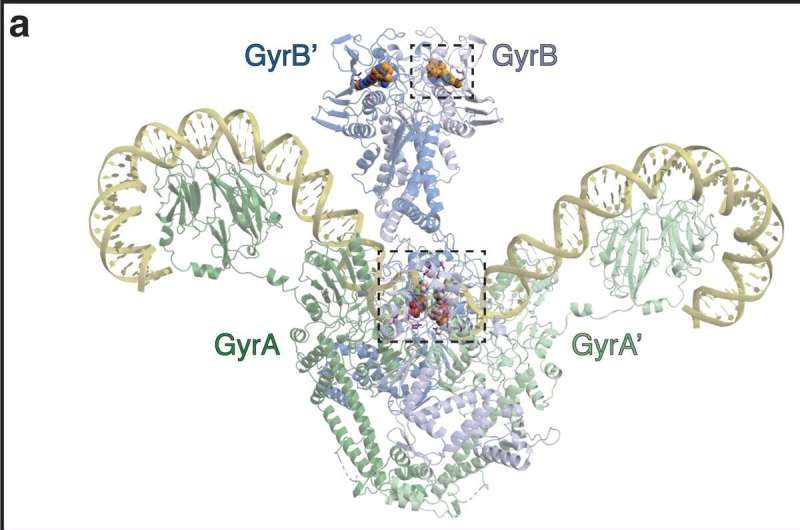
In their paper, Osterman and colleagues combined experimental evolution in a continuous culturing device (morbidostat) with whole genome sequencing of evolving cultures to track how E. coli and A. baumannii acquired drug resistance against a pair of antibiotics that inhibit DNA gyrase, an essential enzyme in bacteria.
Inhibiting the enzyme disrupts DNA synthesis and, subsequently, causes the bacteria to die. One antibiotic—ciprofloxacin—has been in active clinical use since 1987; the other—GP6—is experimental.
The researchers found that pathogenic bacteria with acquired resistance to ciprofloxacin remained susceptible to effective antimicrobial treatment by the GP6 drug. However, the opposite was not true: the evolution of resistance to GP6 also triggered resistance to ciprofloxacin.
These findings underscored that E. coli and A. baumannii employ shared and unique mechanisms to acquire resistance to these two types of drugs.
"Bacteria acquire resistance as a result of random mutational events that happen in the DNA replication as uncorrected 'typos'," said Osterman. "These spontaneously emerge in a handful of drug-resistant variants/strains out of godzillions of neutral mutations) under selective pressure." (A godzillion is a descriptive term, not a precise unit of measurement. It refers to a number of enormous sizes. In this case a number ranging between 108 to 109 neutral mutations.)
These harmful mutations may underlie several types of resistance mechanisms, including modifying the protein targets of a given drug, the ability of bacterial cells to expel compounds (drugs) before they cause harm (efflux), and special bacterial enzymes that inactivate active drug compounds.
Osterman said the work advances progress toward developing "resistibility profiles" of established and novel antimicrobial drugs, which would help set forth "boundaries for possible combinational treatment, including clinically relevant multidrug-resistant strains. Our acquired knowledge provides crucial guidelines for all these translational activities."
The findings are not limited to E. coli and A. baumannii. They have the potential to at least partially predict resistance drivers across other closely related species posing significant health risks, including Pseudomonas, Salmonella, and Klebsiella spp., all difficult-to-treat bacterial pathogens linked to serious and sometimes deadly infections.
More information: Semen A. Leyn et al, Two classes of DNA gyrase inhibitors elicit distinct evolutionary trajectories toward resistance in gram-negative pathogens, npj Antimicrobials and Resistance (2024). DOI: 10.1038/s44259-024-00021-y
Provided by Sanford-Burnham Prebys