This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Researchers investigate archaea to discover how proteins determine cell shape and function
Originally discovered in extreme environments such as hydrothermal vents, archaea, a single-celled microorganism, can also be found in the digestive systems of animals, including humans in which they play a key role in gut health. Yet, little is known about the function of these cells or how they form the distinct shapes they assume to match their environments.
Now, research led by Mecky Pohlschröder of the University of Pennsylvania's School of Arts & Sciences has uncovered key insights into the molecular machinery that determines archaea's morphology. The findings are published in the journal Nature Communications.
"It's always exciting when collaborative research across multiple diverse disciplines culminates in a significant discovery, but it's that much more satisfying when the research deepens our understanding of a fundamental biological process," Pohlschröder says.
"Characterization of the proteins that determine cell shape in archaea may also shed light on the mechanisms underlying other cellular processes in archaea." Pohlschröder notes their findings also point to better understanding such mechanisms in bacteria and eukaryotes, organisms whose cells contain a nucleus within a membrane.
From high school project to cutting-edge research
Lead author Heather Schiller, a former graduate student in Pohlschröder's lab, recounts how this discovery started with a high school student's summer research in the lab. "Joshua, our summer research assistant, was given the laborious task of sampling upwards of 1,000 strains from a library of random mutations to observe their ability to swim," Schiller says. "In doing so, he identified a couple of hyper-motile Haloferax volcanii mutants."
Schiller explains the process involved using agar plates with a consistency that allowed the cells to swim through it, creating visible halos. The mutants that couldn't swim would not form these halos, enabling their identification. The student was looking for mutants that showed altered motility, and among these some moved faster than usual.
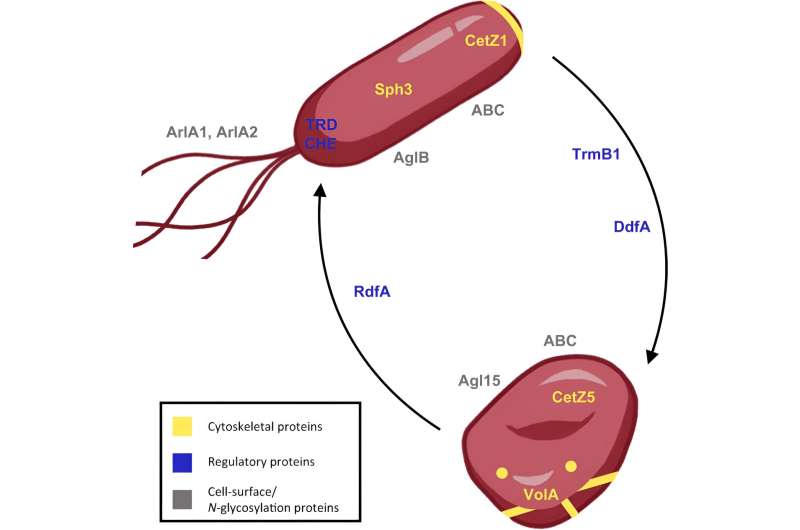
"Upon closer examination under the microscope, these faster-moving mutants were found to only form rod shapes, never disks," Schiller says. "This discovery was crucial for our latest research as it provided a clear direction for further investigation into the cellular components involved in shape formation, beyond just the mutant protein identified."
The importance of rod and disk shapes in Haloferax volcanii is not just a matter of morphology but is intricately linked to the organism's functionality and adaptability to environmental conditions, Pohlschröder explains. Rod shapes are particularly crucial for effective swimming, enabling the microbes to navigate their environments efficiently.
The latest research dives deeper into these shape transitions by employing a holistic and collaborative approach that integrates transposon insertion screens, quantitative proteomics, reverse genetics, and advanced microscopy.
The multipronged approach
The identification of mutant strains that are locked in a specific cell shape enabled the next step in the researcher's analysis pipeline: comparative quantitative proteomics, says Stefan Schulze, a former postdoctoral researcher in the Pohlshröder Lab, now an assistant professor at the Rochester Institute of Technology.
This technique essentially quantifies the abundance of proteins in cells under various conditions, allowing the researchers to compare the protein profiles of the mutants with those of wild-type cells across different growth phases. This comparison helped discern proteins whose abundance changes are specifically associated with cell shape from those related to growth phases.
To confirm the importance of these cell shape associated proteins, Schiller used a reverse genetics approach, wherein the function of a gene of interest is found by observing the effects of its modification or deletion.
"This approach was used to study the roles of specific proteins identified in the proteomics experiment, like disk-determining factor A and rod-determining factor A in cell-shape determination," Schulze says. It also included a protein that Daniel Safer from Penn's Perelman School of Medicine—who has an extensive background with actin proteins in eukaryotes—had at around the same time pointed out as a potential actin homolog but, unlike other actins involved in rod formation, was more abundant in disks.
While proteomics and reverse genetics can determine which proteins are crucial for shifting cell shape, advanced microscopy was needed to visualize the behavior of these proteins inside the cell. A collaboration with Alex Bisson, an assistant professor at Brandeis University, provided the last missing piece to the puzzle. The Bisson lab leveraged expertise in super-resolution microscopy and computational analysis to visualize and quantify the dynamic behavior of proteins like volactin, an actin homolog the researchers identified as crucial in the transition to disk-shaped cells.
Pohlschröder teamed with the Bisson lab to create 3D images of volactin polymers in high resolution inside cells. Using volactin fused to the fluorescent protein GFP, the researchers were able to follow volactin dynamics in real time which showed these polymers elongating (polymerization) and shrinking (depolymerization), suggesting that volactin polymers could dynamically "sense" the cell state in order to coordinate cell shape.
"Unlike its function in many eukaryotic cells, where actin is often associated with maintaining cell shape and facilitating movement, in our model archaeon Haloferax volcanii this actin homolog is crucial for the transition to disk-shaped cells," Pohlschröder says.
Looking ahead
"This research truly underscores the importance of interdisciplinary collaboration and expertise in advancing science across all levels," Pohlschröder says. "Daniel's input, combined with our team's comprehensive approach, with significant contributions from Stefan and Alex, led to such noteworthy insights into the molecular mechanisms of cell-shape determination in archaea, particularly the role of volactin."
Pohlschröder says that, next to further investigating the functions of proteins identified in these studies, she and her team are now interested in finding out how archaea know when to switch from a rod to a disk. She is currently working with researchers in the Department of Chemistry to elucidate signaling molecules that initiate the shift in cell shape.
More information: Heather Schiller et al, Identification of structural and regulatory cell-shape determinants in Haloferax volcanii, Nature Communications (2024). DOI: 10.1038/s41467-024-45196-0
Provided by University of Pennsylvania