This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Genetic variants identified in miracle fruit pulp transcriptomes
Miracle fruit (Synsepalum dulcificum) is the botanical source of miraculin, a natural, noncaloric sweetener. Miracle fruit (Synsepalum dulcificum) is native to West Africa, where it has been used for more than 100 years to increase the palatability of local foods.
Miracle fruit plants exhibit a bush-like architecture and yield multiple flushes of attractive red berries each year. These berries comprise a large seed, opaque pulp, and a brilliant red peel. Within the fruit's pulp resides a glycoprotein called miraculin, which binds to the sweet receptors on the tongue.
When exposed to acidic stimuli, miraculin undergoes a conformational change, resulting in a strong sweet sensation, even in the absence of actual sugars. The popularity of miracle fruit stems from its intriguing taste-modifying properties.
Historically, miracle fruit, like many other tropical species, has lacked foundational molecular, genetic, and genomics information to fully leverage the diversity of this species and create novel, more profitable cultivars. Recently, however, excellent resources have been published for this species. The chloroplast genome of 1.6 Mb with a total of 133 genes, including 88 protein-coding genes, is publicly available.
A miracle fruit chromosome-level genome assembly of ∼550 Mb containing 37,911 genes, along with transcriptomes of various miracle fruit tissues, is also available. Nevertheless, most molecular miracle fruit studies have focused on the miraculin protein rather than genetics or diversity analyses.
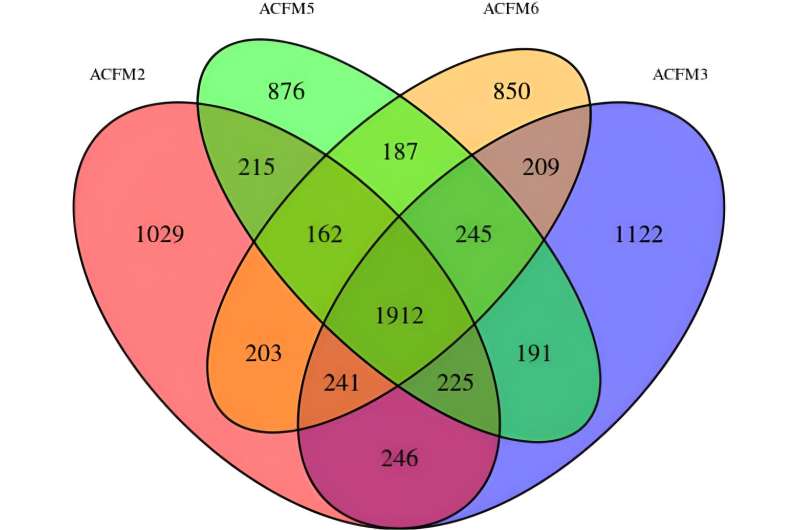
Recent research reports miracle fruit pulp transcriptomes from "Sangria," "Vermilion," "Flame," and "Cherry" morphotypes. A consensus transcriptome included 91,856 transcripts. Reads mapping to the miraculin gene had the highest representation in individual miracle fruit pulp transcriptomes. Other abundant transcripts primarily included Gene Ontology categories representing cellular components, nucleus and nucleic acid binding, and protein modification.
The transcriptomes were used to design real-time quantitative reverse-transcription polymerase chain reaction (qRT-PCR) primers for actin, elongation factor 1α, and the miraculin gene. Analysis by qRT-PCR indicated that miracle fruit pulp and peel tissues had the highest abundance of miraculin transcripts, although other tissues such as leaf, root, and flower also had detectable levels of the target sequence.
Overall, these results will support discovery research for miracle fruit and the eventual breeding of this species.
Advancements in miracle fruit genomics and interest in plant breeding of this species are encouraging. The development of SNP markers will benefit many studies of this species. The unique SNP sets identified during this study could be used to genotype miracle fruit collections previously characterized for fruit quality and yield.
This would help resolve previous challenges arising from unknown genetics that complicate the grouping of morphotypes for data analysis. The identified SNPs could also be used to create low-cost genotyping assays for diversity collections in Africa. Additionally, SNPs located in exons could have biological implications that are worth pursuing in the future.
Miracle fruit is an intriguing species that has benefited from researcher interest. The genomics work performed during this study will benefit future studies aimed at the genetic improvement of this species. Future economic opportunities for miracle fruit could depend on genetic advancements of this species.
The work is published in the Journal of the American Society for Horticultural Science.
More information: Vincent Njung'e Michael et al, Miracle Fruit Pulp Transcriptomes Identify Genetic Variants in Support of Discovery Research and Breeding, Journal of the American Society for Horticultural Science (2023). DOI: 10.21273/JASHS05312-23
Journal information: Journal of the American Society for Horticultural Science
Provided by American Society for Horticultural Science